| (1) HEART | (1) KIDNEY | (2) TESTES |
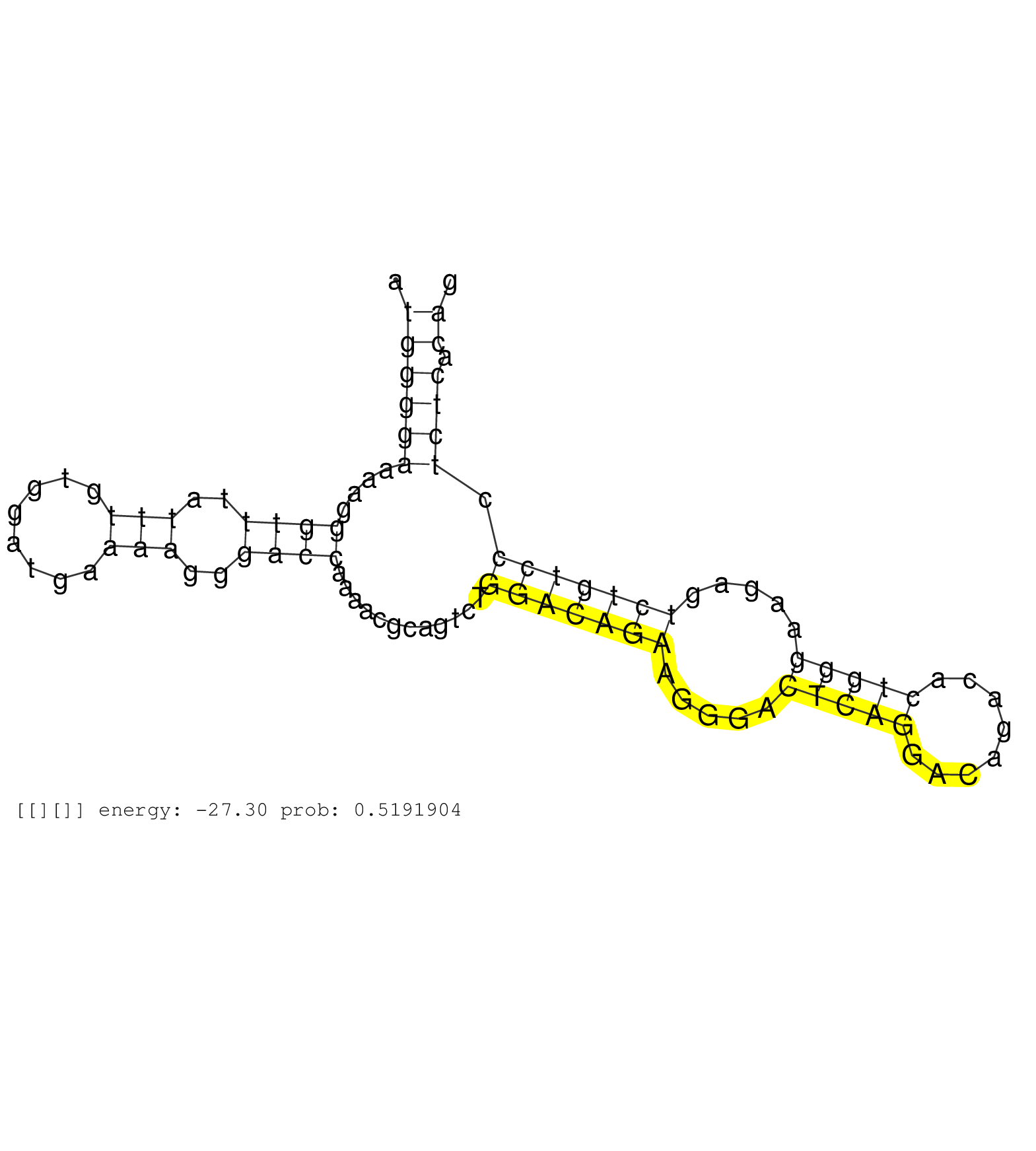
| CATCCGGAGGCCAGGAGGCCTGGGTGTGCCTTGCCAGAGAAGTGCAGGCGGGTAGTAAAGGGGGCCTCCCTCCTGCAACTGCTGAGCAGCCTCCACGTGCATCCTTTGTGGGGAGGAACAAGCCCCAAACGCAGACCCTCGTGTGTATAAAACACAAGAAGATCTCCCGAGGGAGGCCGAGGGTCCTACACAGCCCAGAGATGGGGAAAAGGGTTTATTTGTGGATGAAAAGGGACCAAAACGCAGTCTGGACAGAAGGGACTCAGGACAGACACTGGGAAGAGTCTGTCCCTCTCACAGGTGCACTATGCAGCTGGAAACGACAACAGGCCTGCCCTGGTGCCGATGGC .........................................................................................................................................................................................................((((((....((((..(((........)))..))))............(((((((.....(((((........))))).....))))))).)))).))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553579(SRX182785) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................................CAAAACGCAGTCTGGACAGAAGGGAC........................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .................................................................................................................................................................................................................................TGAAAAGGGACCAAAACGCAGTCTGGACAG............................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ........................................................................................................................................................................................................................................................TGGACAGAAGGGACTCAGGACAGACACTGG........................................................................ | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ........................................................................................................................................................................................................................................................TGGACAGAAGGGACTCAGGAC................................................................................. | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .......................................................................................................................................................................................................................TATTTGTGGATGAAAAGGGACCAAAACGCA......................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .........................................................................TGCAACTGCTGAGCAGCCTCCACGTG........................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .........................................................................................................................................................................................................................TTTGTGGATGAAAAGGGACCAAAACGC.......................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ........................................................................................................................................................................................................................................................TGGACAGAAGGGACTCAGGACAGACAC........................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................................................................GAAGGGACTCAGGACA................................................................................ | 16 | 5 | 0.20 | 0.20 | - | - | - | 0.20 |
| CATCCGGAGGCCAGGAGGCCTGGGTGTGCCTTGCCAGAGAAGTGCAGGCGGGTAGTAAAGGGGGCCTCCCTCCTGCAACTGCTGAGCAGCCTCCACGTGCATCCTTTGTGGGGAGGAACAAGCCCCAAACGCAGACCCTCGTGTGTATAAAACACAAGAAGATCTCCCGAGGGAGGCCGAGGGTCCTACACAGCCCAGAGATGGGGAAAAGGGTTTATTTGTGGATGAAAAGGGACCAAAACGCAGTCTGGACAGAAGGGACTCAGGACAGACACTGGGAAGAGTCTGTCCCTCTCACAGGTGCACTATGCAGCTGGAAACGACAACAGGCCTGCCCTGGTGCCGATGGC .........................................................................................................................................................................................................((((((....((((..(((........)))..))))............(((((((.....(((((........))))).....))))))).)))).))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553579(SRX182785) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................caagGGAGATCTTCTGAAC................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | 1.00 | - |