| (2) TESTES |
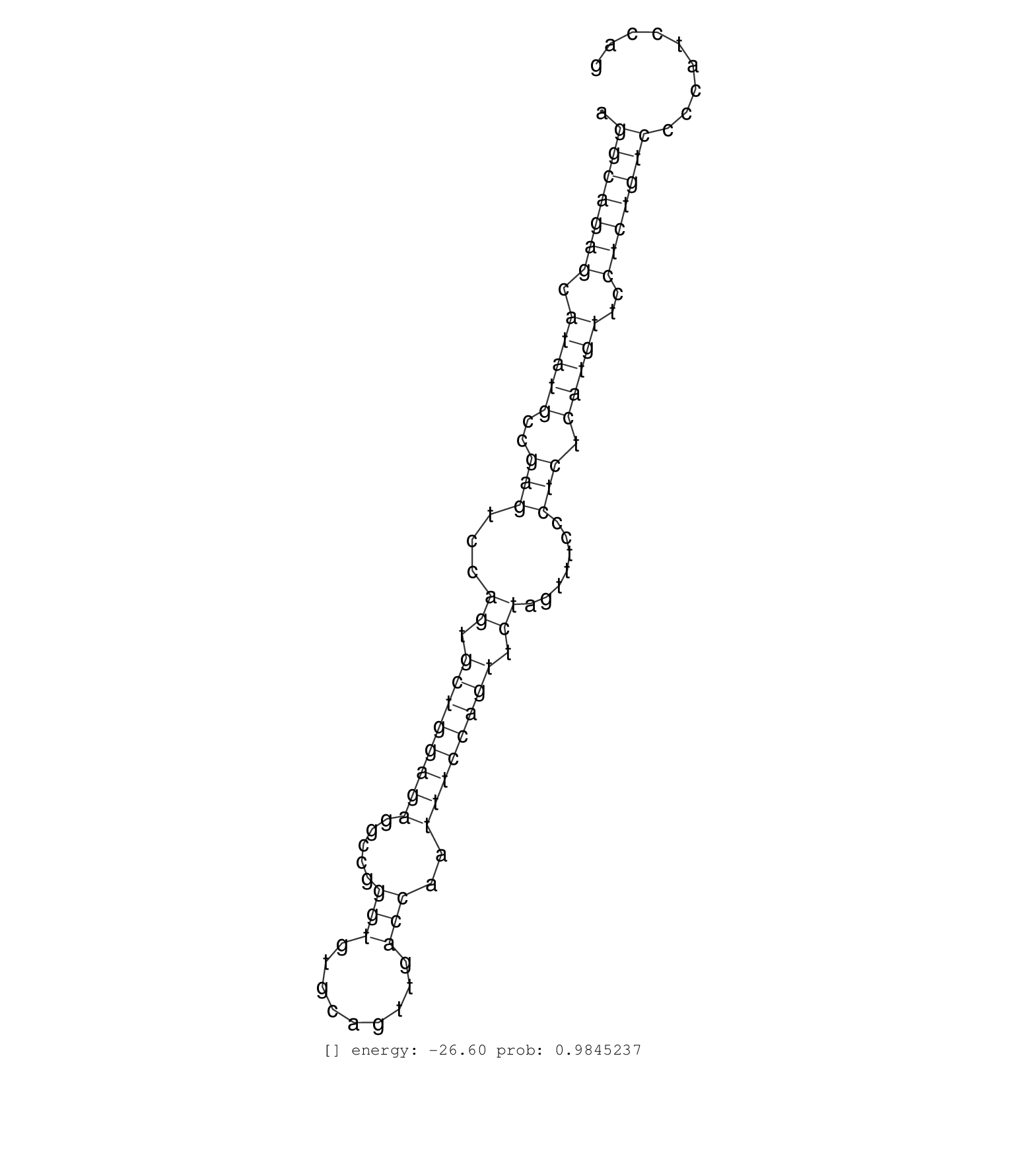
| GATGTCCGGATGCTGGCAATGAGTAGGCCTTCCCTACGCTGGGTGGCGTCCACACCCTTTGGCTTCCATTGCCTGGGTCTCCTGGAGGTGATTTGCTGGATGAATACTGCATGCACAGAATCTGGCCTTGGGTTTGAATGTGGCGGCCAGTGGACAGCATGTGCTTCAGTTATGAGGCTGCCCAGGAGATGCTTCTTCCAAGGCAGAGCATATGCCGAGTCCAGTGCTGGAGAGGCCGGGTGTGCAGTTGACCAATTTCCAGTTCTAGTTTCCCTCTCATGTTCCTCTGTCCCCATCCAGGACATGCTCTGGAGTCAGAAGACAGCAAAAAGAGAAGCAGAAGTCCCGGT .........................................................................................................................................................................................................(((((((.(((((..(((...((.((((((((.....(((.........)))..)))))))).)).......))).)))))..)))))))........................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553607(SRX182813) source: Testis. (testes) | SRR553581(SRX182787) source: Testis. (testes) |
|---|---|---|---|---|---|---|
| ............................................................................................TTGCTGGATGAATACTGCATGCACAGAATC.................................................................................................................................................................................................................................... | 30 | 1 | 10.00 | 10.00 | 5.00 | 5.00 |
| ................................................................................................TGGATGAATACTGCATGCACAGAATCTGGC................................................................................................................................................................................................................................ | 30 | 1 | 4.00 | 4.00 | 4.00 | - |
| ................................................................................................................................................................................................................................................................................................................TGCTCTGGAGTCAGAAGACAGCAAAAAGAGA............... | 31 | 1 | 2.00 | 2.00 | - | 2.00 |
| ...................................................................................................................................................................................................................................................................................................................TCTGGAGTCAGAAGACAGCAAAAAGAGAAGC............ | 31 | 1 | 2.00 | 2.00 | 2.00 | - |
| ............................................................................................................................................................................................................................................................................................................GACATGCTCTGGAGTCAGAAGACAGCAA...................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - |
| ............................................................................................................................................................................................................................................................................................................GACATGCTCTGGAGTCAGAAGACAGCAAAA.................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - |
| ................................................................................................................................................................................................................................................................................................................TGCTCTGGAGTCAGAAGACAGCAAAAAGAGAA.............. | 32 | 1 | 1.00 | 1.00 | - | 1.00 |
| ...................................................................................................................................................................................................................................................................................................................TCTGGAGTCAGAAGACAGCAAAAAGAGA............... | 28 | 1 | 1.00 | 1.00 | - | 1.00 |
| ................................................................................................................................................................................................................................................................................................................TGCTCTGGAGTCAGAAGACAGCAAAAAGAGT............... | 31 | 1.00 | 0.00 | - | 1.00 | |
| ................................................................................................................................................................................................................................................................................................................TGCTCTGGAGTCAGAAGACAGCAAAAAGATA............... | 31 | 1.00 | 0.00 | - | 1.00 | |
| ..................................................................................................................................................................................................................................................................................................................CTCTGGAGTCAGAAGACAGCAAAAAGAGAAGC............ | 32 | 1 | 1.00 | 1.00 | - | 1.00 |
| ............................................................................................TTGCTGGATGAATACTGCATGCACAGA....................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - |
| GATGTCCGGATGCTGGCAATGAGTAGGCCTTCCCTACGCTGGGTGGCGTCCACACCCTTTGGCTTCCATTGCCTGGGTCTCCTGGAGGTGATTTGCTGGATGAATACTGCATGCACAGAATCTGGCCTTGGGTTTGAATGTGGCGGCCAGTGGACAGCATGTGCTTCAGTTATGAGGCTGCCCAGGAGATGCTTCTTCCAAGGCAGAGCATATGCCGAGTCCAGTGCTGGAGAGGCCGGGTGTGCAGTTGACCAATTTCCAGTTCTAGTTTCCCTCTCATGTTCCTCTGTCCCCATCCAGGACATGCTCTGGAGTCAGAAGACAGCAAAAAGAGAAGCAGAAGTCCCGGT .........................................................................................................................................................................................................(((((((.(((((..(((...((.((((((((.....(((.........)))..)))))))).)).......))).)))))..)))))))........................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553607(SRX182813) source: Testis. (testes) | SRR553581(SRX182787) source: Testis. (testes) |
|---|