| (1) HEART | (2) TESTES |
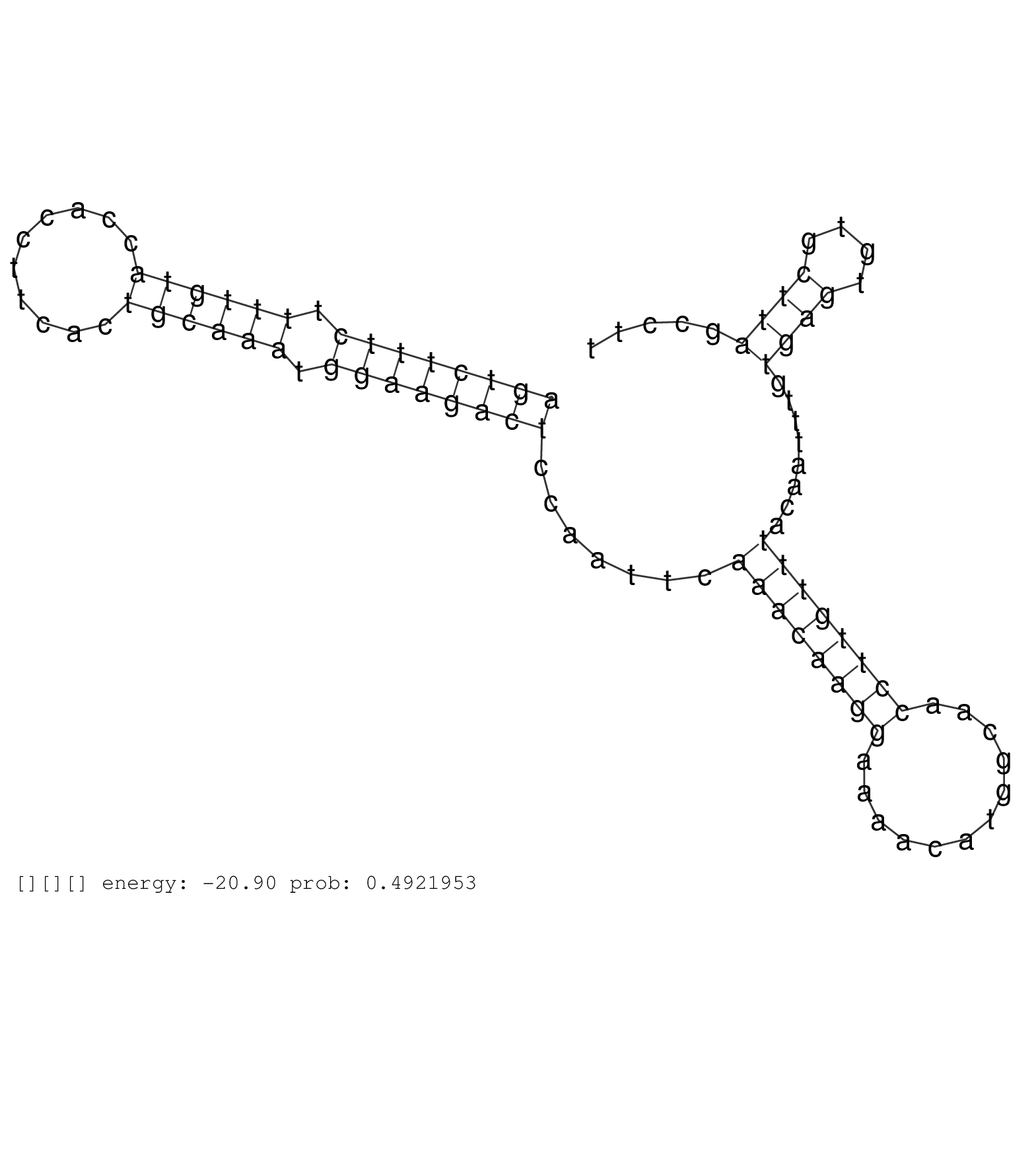
| ACAGTGCCCTCAGTGACTCCATCCACACTGTGGCCATGAGCACCAACTCTGTAAGCGTGGCACTCTCTACCTCACACAACCTTGCCTCCCTAGAGTCTGTTTCCCTCCATGAAGTTGGCCTCAGCCTAGAACCTGTGGCTGTCTCCTCCATCACCCAGGAGGTTGCTATGGGGACAGGTCATGTAGATGTATCTTCAGACAGTCTTTCTTTTGTACCACCTTCACTGCAAATGGAAGACTCCAATTCAAACAAGGAAAACATGGCAACCTTGTTTACAATTTGTGAGTGTGCTTAGCCTTTCTTTTTTGGAAATATTTGTAAAAAGGGCCACCATCCCTAACCAGTTAAG ........................................................................................................................................................................................................((((((((.((((((..........)))))).)))))))).......((((((((............))))))))........((((....))))....................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553579(SRX182785) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................TTGTACCACCTTCACTGCAAATGGAAGACTC............................................................................................................. | 31 | 1 | 2.00 | 2.00 | - | 2.00 | - |
| ..........................................................................................................................................................................GGGACAGGTCATGTAGATGTATCTTCAGA....................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 2.00 | - | - |
| ..........................................................................................................................................................................GGGACAGGTCATGTAGATGTATCTTCAGACA..................................................................................................................................................... | 31 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - |
| ..............................TGGCCATGAGCACCAACTCTGTAAGCGTGGC................................................................................................................................................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ..........................................................................................TAGAGTCTGTTTCCCTCCATGAAGTT.......................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ........................................CACCAACTCTGTAAGCGTGGCACTCTCTA......................................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ..............................................................................................................................TAGAACCTGTGGCTGTC............................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ...............................................................................................TCTGTTTCCCTCCATGAAGTTGGCCTCAGCC................................................................................................................................................................................................................................ | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .................................................TGTAAGCGTGGCACTCTCTACCTCACACAACA............................................................................................................................................................................................................................................................................. | 32 | 1.00 | 0.00 | - | 1.00 | - | |
| ..............................................................................................................................TAGAACCTGTGGCTGTCTCCTCCATCACCC.................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...............................................TCTGTAAGCGTGGCACTCTCTACCTCACA.................................................................................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .....................................................................................................................................................................................................................TACCACCTTCACTGCAAATGGAAGACTCC............................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...............................................................................................................................................................................................TCTTCAGACAGTCTTTCTTTTGTACCACCT................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...............................................................................................................................................................................AGGTCATGTAGATGTATCTTCAGACA..................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ACAGTGCCCTCAGTGACTCCATCCACACTGTGGCCATGAGCACCAACTCTGTAAGCGTGGCACTCTCTACCTCACACAACCTTGCCTCCCTAGAGTCTGTTTCCCTCCATGAAGTTGGCCTCAGCCTAGAACCTGTGGCTGTCTCCTCCATCACCCAGGAGGTTGCTATGGGGACAGGTCATGTAGATGTATCTTCAGACAGTCTTTCTTTTGTACCACCTTCACTGCAAATGGAAGACTCCAATTCAAACAAGGAAAACATGGCAACCTTGTTTACAATTTGTGAGTGTGCTTAGCCTTTCTTTTTTGGAAATATTTGTAAAAAGGGCCACCATCCCTAACCAGTTAAG ........................................................................................................................................................................................................((((((((.((((((..........)))))).)))))))).......((((((((............))))))))........((((....))))....................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553579(SRX182785) source: Heart. (Heart) |
|---|