| (1) AGO1.ip | (4) AGO2.ip | (2) AGO3.ip | (2) B-CELL | (2) BRAIN | (19) BREAST | (41) CELL-LINE | (5) CERVIX | (4) HEART | (3) HELA | (7) LIVER | (3) OTHER | (40) SKIN | (1) TESTES | (1) UTERUS | (1) XRN.ip |
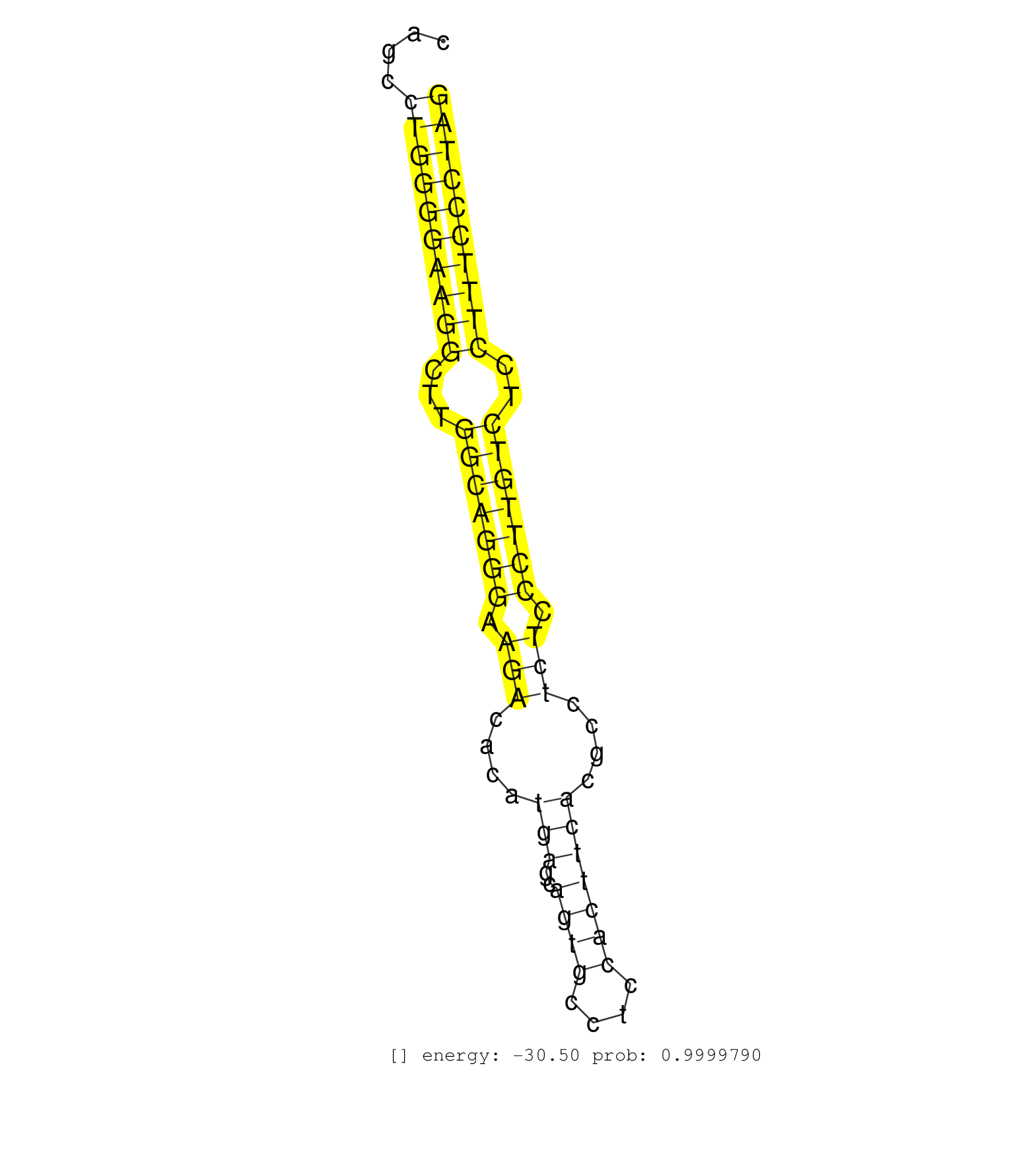
| AACCCAGGGCTGAAAAGGATTTGAAGGGCAGGATTTCCAGAAAGTACTGAGGTGAGGAAGAAAACCAAGACTAGGGCAGTGCAGAGTATCTGGGAGCTTAGAGTACTGAACATTCTGCCAGCAGCCTGGGGAAGGCTTGGCAGGGAAGACACATGAGCAGTGCCTCCACTTCACGCCTCTCCCTTGTCTCCTTTCCCTAGCTTGTGACAGAGCTGCTGAGTGGCATTCGGGTCATCAAGTTCTGCGGGTG .............................................................................................................................((((((((((...(((((((.(((....(((..((((....)))))))....))).)))))))..)))))))))).................................................. .........................................................................................................................122...........................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189782 | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | TAX577743(Rovira) total RNA. (breast) | SRR343334 | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189784 | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191621(GSM715731) 16genomic small RNA (size selected RNA from t. (breast) | SRR553576(SRX182782) source: Testis. (testes) | SRR029131(GSM416760) MCF7. (cell line) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | TAX577580(Rovira) total RNA. (breast) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | GSM956925Ago2PAZ(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR040031(GSM532916) G013T. (cervix) | SRR191592(GSM715702) 59genomic small RNA (size selected RNA from t. (breast) | SRR040028(GSM532913) G026N. (cervix) | SRR038858(GSM458541) MEL202. (cell line) | SRR189786 | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR040011(GSM532896) G529T. (cervix) | SRR191570(GSM715680) 56genomic small RNA (size selected RNA from t. (breast) | SRR189783 | SRR191422(GSM715532) 129genomic small RNA (size selected RNA from . (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR040016(GSM532901) G645N. (cervix) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR191454(GSM715564) 180genomic small RNA (size selected RNA from . (breast) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) | SRR191612(GSM715722) 65genomic small RNA (size selected RNA from t. (breast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR191492(GSM715602) 154genomic small RNA (size selected RNA from . (breast) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038856(GSM458539) D11. (cell line) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | GSM532874(GSM532874) G699T. (cervix) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR390723(GSM850202) total small RNA. (cell line) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | GSM956925Paz8D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | DRR000560(DRX000318) Isolation of RNA following immunoprecipitatio. (ago1 cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR191603(GSM715713) 71genomic small RNA (size selected RNA from t. (breast) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR191554(GSM715664) 99genomic small RNA (size selected RNA from t. (breast) | SRR191629(GSM715739) 5genomic small RNA (size selected RNA from to. (breast) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR191537(GSM715647) 45genomic small RNA (size selected RNA from t. (breast) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR343335 | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR029128(GSM416757) H520. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCCTAGT................................................. | 22 | 1 | 84.00 | 34.00 | - | 3.00 | 8.00 | 7.00 | 2.00 | 4.00 | - | 4.00 | 4.00 | 1.00 | - | - | 1.00 | - | - | 2.00 | 1.00 | 3.00 | 2.00 | - | 1.00 | 2.00 | - | - | - | - | 1.00 | 2.00 | - | 2.00 | - | 1.00 | 2.00 | - | 1.00 | 2.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - |
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCCTAG.................................................. | 21 | 1 | 34.00 | 34.00 | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | 1.00 | - | 1.00 | 2.00 | - | 1.00 | - | 1.00 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCCTAGA................................................. | 22 | 1 | 22.00 | 34.00 | 4.00 | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGGGAAGGCTTGGCAGGGAAGA..................................................................................................... | 23 | 1 | 11.00 | 11.00 | 6.00 | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................TGGGGAAGGCTTGGCAGGGAAG...................................................................................................... | 22 | 1 | 11.00 | 11.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CTGGGGAAGGCTTGGCAGGGAAGA..................................................................................................... | 24 | 1 | 9.00 | 9.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCCTATT................................................. | 22 | 1 | 7.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CCTGGGGAAGGCTTGGCAGGGAAGA..................................................................................................... | 25 | 1 | 5.00 | 5.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................CTGGGGAAGGCTTGGCAGGGA........................................................................................................ | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGGGAAGGCTTGGCAGGGAAA...................................................................................................... | 22 | 1 | 4.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CCTGGGGAAGGCTTGGCAGGGAAT...................................................................................................... | 24 | 1 | 4.00 | 1.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCCTAT.................................................. | 21 | 1 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CCTGGGGAAGGCTTGGCAGGGATT...................................................................................................... | 24 | 1 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CCTGGGGAAGGCTTGGCAGGGAAG...................................................................................................... | 24 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................GAAAGTACTGAGGTGAGGAAGA............................................................................................................................................................................................. | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGGGAAGGCTTGGCAGGGAAT...................................................................................................... | 22 | 1 | 3.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGGGAAGGCTTGGCAGGGAA....................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CCTGGGGAAGGCTTGGCAGGGAAGAT.................................................................................................... | 26 | 1 | 2.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CTGGGGAAGGCTTGGCAGGGAA....................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................CTGGGGAAGGCTTGGCAGGGAAGAT.................................................................................................... | 25 | 1 | 2.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CCTGGGGAAGGCTTGGCAGGGA........................................................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCCT.................................................... | 19 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGGGAAGGCTTGGCAGGGAAGAA.................................................................................................... | 24 | 1 | 2.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGGGAAGGCTTGGCAGGGA........................................................................................................ | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CTGGGGAAGGCTTGGCAGGGAAG...................................................................................................... | 23 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGGATTTGAAGGGCAGGATTTCCAG.................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................CTTGTGACAGAGCTGCTGAGTGGCATTCGGG................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCCTAGTAGA.............................................. | 25 | 1 | 1.00 | 34.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGGGAAGGCTTGGCAGGGAAGAC.................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................ACAGAGCTGCTGAGTGGCATTCGGGTCAT............... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGGGAAGGCTTGGCAGGGAT....................................................................................................... | 21 | 1 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CTGGGGAAGGCTTGGCAGGGAAGGT.................................................................................................... | 25 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TTGTGACAGAGCTGCTGAGTGGCATTC...................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCCTTAGA................................................ | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCCTAAAA................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................GGGGAAGGCTTGGCAGGGAA....................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CTGGGGAAGGCTTGGCAGGGAAA...................................................................................................... | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TTGTGACAGAGCTGCTGAGTGGCA......................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................ACAGAGCTGCTGAGTGGCATC....................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................CTGGGGAAGGCTTGGCAGGGAAT...................................................................................................... | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CCTGGGGAAGGCTTGGCAGG.......................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGACAGAGCTGCTGAGTGGC.......................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................CAGGATTTCCAGAAAATA............................................................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCCTA................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTCCCTTGTCTCCTTTCCCTAG.................................................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGGGAAGGCTTGGCAGGGAAGT..................................................................................................... | 23 | 1 | 1.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............AGGATTTGAAGGGCAGTGCT....................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCCTATTT................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CTTGGCAGGGAAGACACATGAGC............................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGGGAAGGCTTGGCAGGGAAGTT.................................................................................................... | 24 | 1 | 1.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................CTGGGGAAGGCTTGGCAGGAAAA...................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGGGAAGGCTTGGCAGGGATA...................................................................................................... | 22 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCCTAGTTT............................................... | 24 | 1 | 1.00 | 34.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....AGGGCTGAAAAGGATTTGAAGG............................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................ACTAGGGCAGTGCAGAGGAGG................................................................................................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................TGTGACAGAGCTGCTGAGTGGCATTC...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGTGACAGAGCTGCTGAGTGGCAT........................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CCTGGGGAAGGCTTGGCAGGGAA....................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........CTGAAAAGGATTTGAAGATA............................................................................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..CCCAGGGCTGAAAAGGATTTGAAGG............................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGGGGAAGGCTTGGCAGGGGAAA..................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................GCCTGGGGAAGGCTTGGCAGGGGAGA..................................................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................TAGGGCAGTGCAGAGGTT................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................CTGGGGAAGGCTTGGCAGG.......................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCCTAGTT................................................ | 23 | 1 | 1.00 | 34.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CCTGGGGAAGGCTTGGCAG........................................................................................................... | 19 | 2 | 1.00 | 1.00 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCCTTGTCTCCTTTCCATA................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................AGGGCAGTGCAGAGTATC................................................................................................................................................................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| .........................................................................................................................................................................................................TTGTGACAGAGCTGCTGAG.............................. | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ............................................................................................................................CCTGGGGAAGGCTTGGCA............................................................................................................ | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AACCCAGGGCTGAAAAGGATTTGAAGGGCAGGATTTCCAGAAAGTACTGAGGTGAGGAAGAAAACCAAGACTAGGGCAGTGCAGAGTATCTGGGAGCTTAGAGTACTGAACATTCTGCCAGCAGCCTGGGGAAGGCTTGGCAGGGAAGACACATGAGCAGTGCCTCCACTTCACGCCTCTCCCTTGTCTCCTTTCCCTAGCTTGTGACAGAGCTGCTGAGTGGCATTCGGGTCATCAAGTTCTGCGGGTG .............................................................................................................................((((((((((...(((((((.(((....(((..((((....)))))))....))).)))))))..)))))))))).................................................. .........................................................................................................................122...........................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189782 | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | TAX577743(Rovira) total RNA. (breast) | SRR343334 | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189784 | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191621(GSM715731) 16genomic small RNA (size selected RNA from t. (breast) | SRR553576(SRX182782) source: Testis. (testes) | SRR029131(GSM416760) MCF7. (cell line) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | TAX577580(Rovira) total RNA. (breast) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | GSM956925Ago2PAZ(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | DRR000559(DRX000317) "THP-1 whole cell RNA, no treatment". (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR040031(GSM532916) G013T. (cervix) | SRR191592(GSM715702) 59genomic small RNA (size selected RNA from t. (breast) | SRR040028(GSM532913) G026N. (cervix) | SRR038858(GSM458541) MEL202. (cell line) | SRR189786 | SRR095854(SRX039177) "miRNA were isolated from FirstChoice Human B. (brain) | SRR040011(GSM532896) G529T. (cervix) | SRR191570(GSM715680) 56genomic small RNA (size selected RNA from t. (breast) | SRR189783 | SRR191422(GSM715532) 129genomic small RNA (size selected RNA from . (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR040016(GSM532901) G645N. (cervix) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR191454(GSM715564) 180genomic small RNA (size selected RNA from . (breast) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) | SRR191612(GSM715722) 65genomic small RNA (size selected RNA from t. (breast) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR191492(GSM715602) 154genomic small RNA (size selected RNA from . (breast) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038856(GSM458539) D11. (cell line) | SRR330864(SRX091702) tissue: skin psoriatic involveddisease state:. (skin) | GSM532874(GSM532874) G699T. (cervix) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR390723(GSM850202) total small RNA. (cell line) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | GSM956925Paz8D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (cell line) | DRR000560(DRX000318) Isolation of RNA following immunoprecipitatio. (ago1 cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR191603(GSM715713) 71genomic small RNA (size selected RNA from t. (breast) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR191554(GSM715664) 99genomic small RNA (size selected RNA from t. (breast) | SRR191629(GSM715739) 5genomic small RNA (size selected RNA from to. (breast) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR191537(GSM715647) 45genomic small RNA (size selected RNA from t. (breast) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR343335 | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR029128(GSM416757) H520. (cell line) | SRR029125(GSM416754) U2OS. (cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................CAGAGCTGCTGAGTGGCT......................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................................................CAAGTTCTGCGGGTGTC | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CTCCCTTGTCTCCTTTCC...................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CTCTCCCTTGTCTCCTTT........................................................ | 18 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TGTCTCCTTTCCCTAGGGC............................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................GCAGAGTATCTGGGAGCTT....................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GGCAGGATTTCCAGAAAG.............................................................................................................................................................................................................. | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |