Genes highly conserved to chicken (n=85).
This analysis looks at the genes we determined
The base scores are calculated using a weighted measure of conservation for human, chimp, mouse, dog, and chicken.
The target scores use the following method:
Control type: Shuffle
3' UTR:Must be in all species to be included in count
tolerance: 20%
min controls allowed: 3
max controls: 50
Note: See my project wiki for more information on methods.
Alignments
View all genes used in this data set.
View HCMRDC alignments or 28-way alignments
Web logos (click for full-size image)
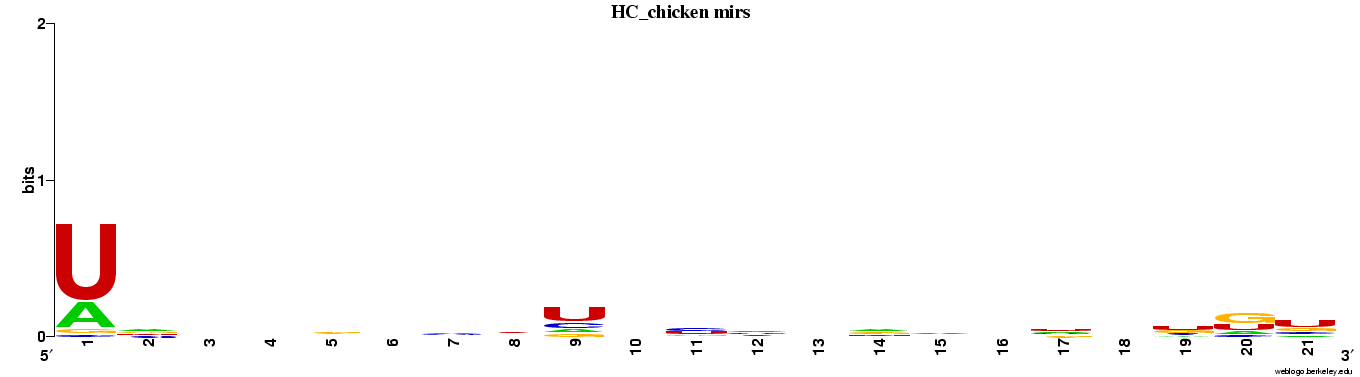
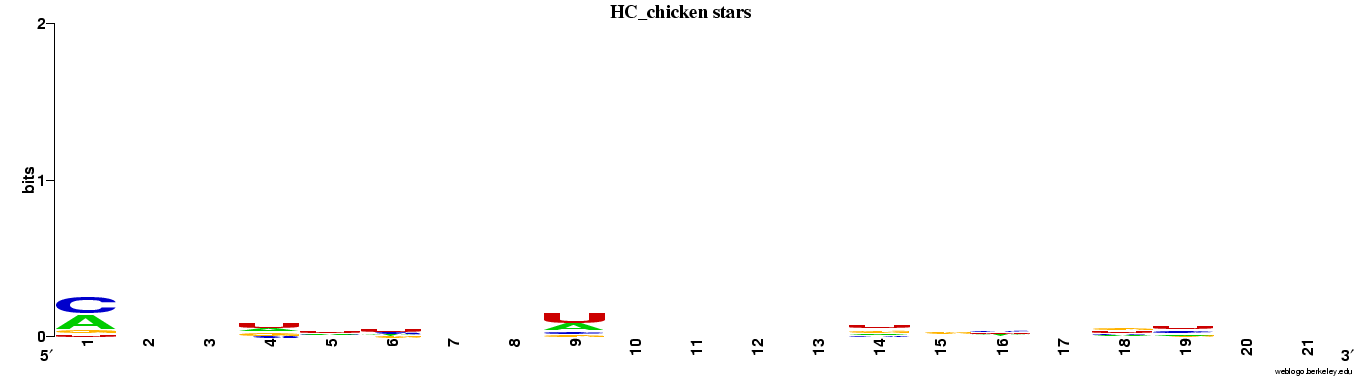
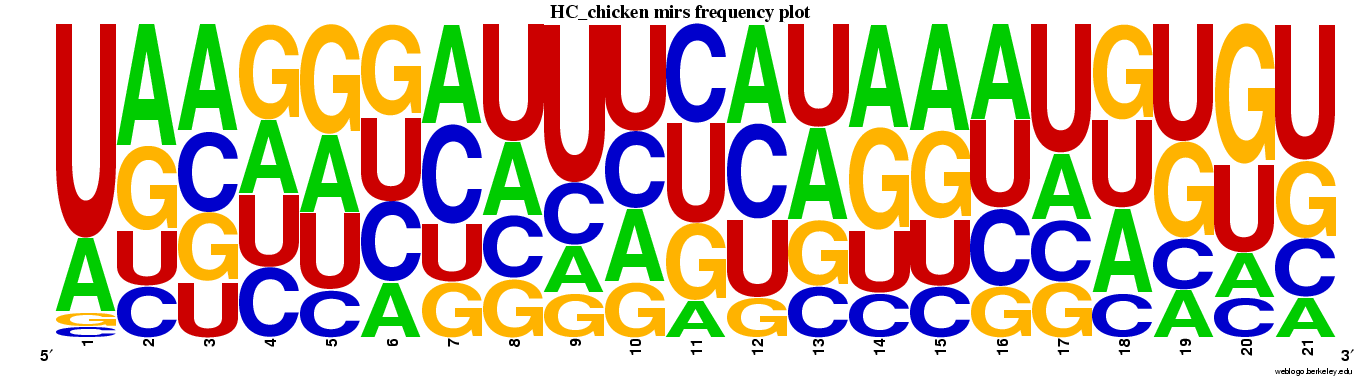
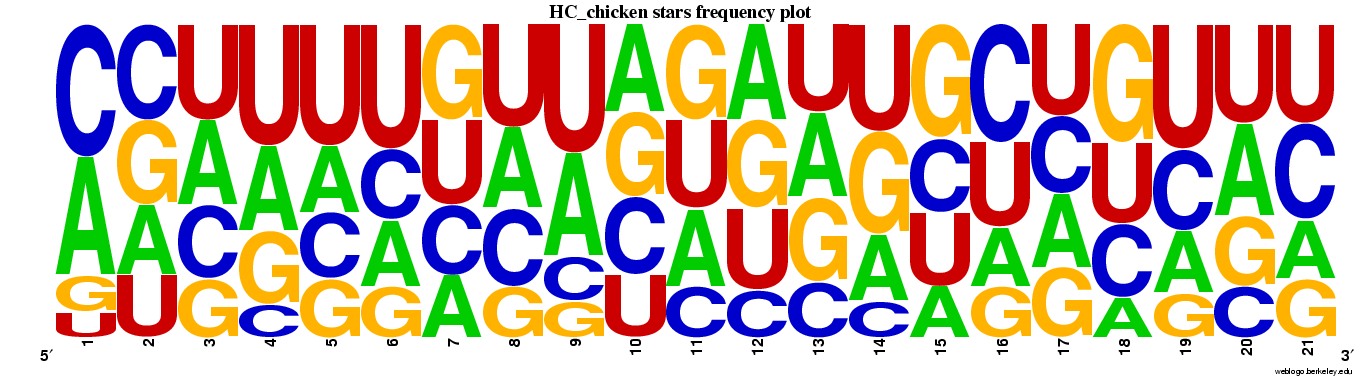
Spreadsheets
Flanking Base Scores: Contains base conservation scores (using HCMDC) for each base in the miR and miR* as well as 7nt of flanking sequence.
download spreadsheet view webpage
Window Scores: Calculates average over 7nt windows using same conservation scores as above.
download spreadsheet view webpage
Target Scores (5-way): Number of conserved targets, signal-to-noise of conserved targets, percentage in each window with s2n > 1, percentage in each window with p-value < .05, z-scores, tests comparing each windows s2n score to background model of all possible 7mers.
Target scores use human, chimp, mouse, rat, and dog (5-way).
download spreadsheet view webpage



