| (1) AGO.mut | (4) AGO2.ip | (1) AGO3.ip | (6) B-CELL | (15) BRAIN | (3) CELL-LINE | (1) DCR.mut | (6) EMBRYO | (6) ESC | (1) FIBROBLAST | (3) LIVER | (1) LUNG | (4) LYMPH | (3) OTHER | (2) OTHER.mut | (6) PIWI.ip | (2) PIWI.mut | (1) SKIN | (2) SPLEEN | (1) TDRD1.ip | (20) TESTES | (1) THYMUS | (1) TOTAL-RNA |
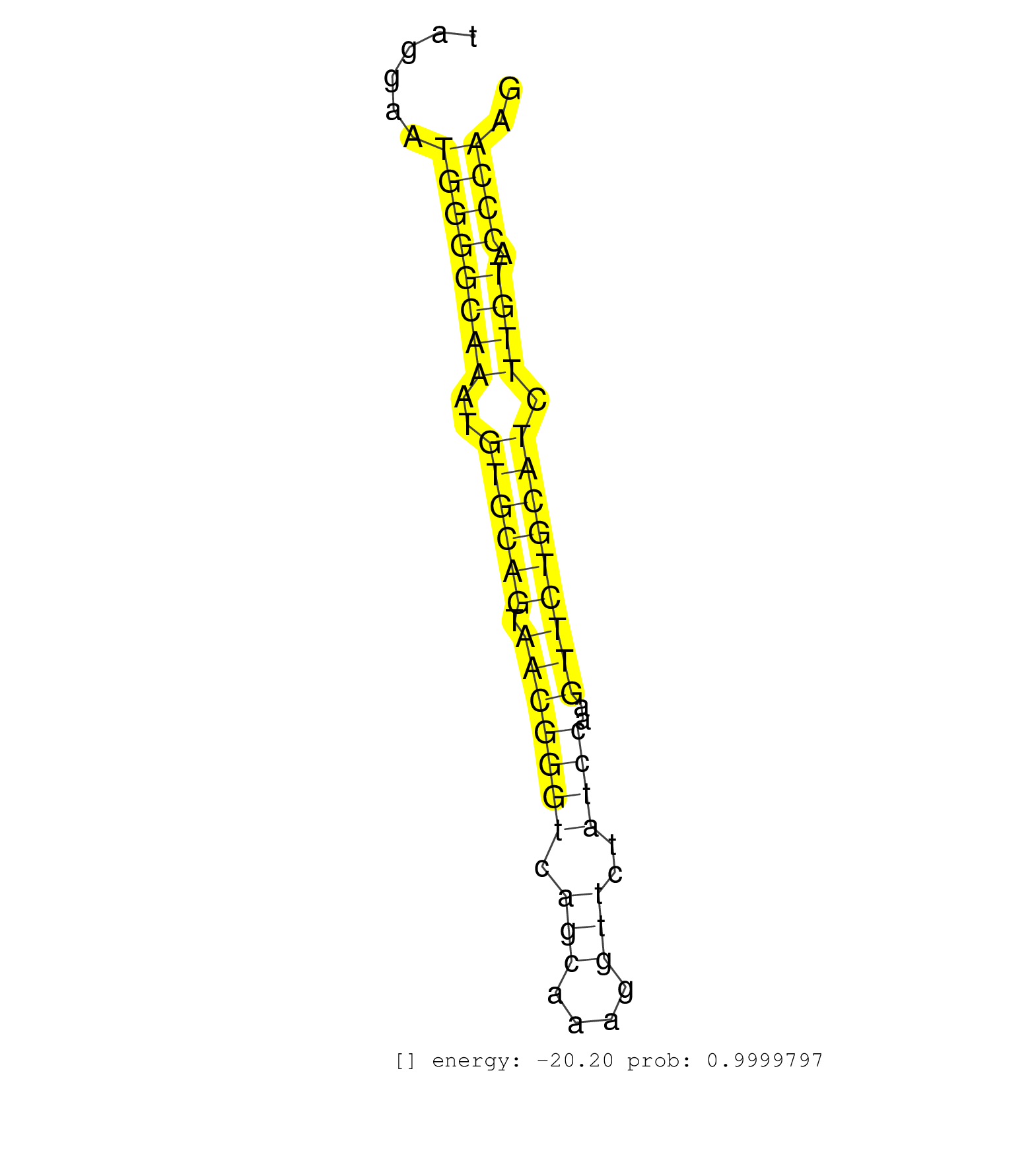
| CCCAGCAGAGAGCACTAGCATTGCAAGTGTTGTCCCAGATCGTCGGCAGGGTAAGTGGGCTGCTGGCCTGGTCCTACCAAGCCACACCCCATCCTTCATCCCTTGATCTTAGCCTTTTCCTCCTTACTGTGGGTCAGACTAGGAATGGGGCAAATGTGCAGTAACGGGTCAGCAAAGGTTCTATCCAAGTTCTGCATCTTGTACCCAAGGCCCAGGCTGGTGAGTTTGGGGACCGCCTAGTGGGCAGTGTCTTGCGCCT .................................................................................................................................................((((((((..((((((.(((((((.(((....)))..))))..))))))))).)))).)))).................................................... ...........................................................................................................................................140..................................................................209................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | mjTestesWT4() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | mjLiverWT2() Liver Data. (liver) | SRR685340(GSM1079784) Small RNAs (15-50 nts in length) from immorta. (dicer cell line) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR553604(SRX182810) source: Spermatocytes. (Spermatocytes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR553582(SRX182788) source: Brain. (Brain) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR059765(GSM562827) DN3_control. (thymus) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | mjLiverWT3() Liver Data. (liver) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR206941(GSM723282) other. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | GSM475281(GSM475281) total RNA. (testes) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR553603(SRX182809) source: Spermatids. (Spermatids) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | GSM510452(GSM510452) newborn_rep8. (total RNA) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................GTTCTGCATCTTGTACCCAAG.................................................. | 21 | 1 | 18.00 | 18.00 | 9.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATGGGGCAAATGTGCAGTAACGGG........................................................................................... | 24 | 1 | 16.00 | 16.00 | - | 7.00 | 2.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................TCCCAGATCGTCGGCAGG................................................................................................................................................................................................................. | 18 | 1 | 10.00 | 10.00 | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................TTGCAAGTGTTGTCCCAGATCGTCGGC.................................................................................................................................................................................................................... | 27 | 1 | 8.00 | 8.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................AGTTCTGCATCTTGTACCCAAG.................................................. | 22 | 1 | 7.00 | 7.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATGGGGCAAATGTGCAGTAACGG............................................................................................ | 23 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................CCTTACTGTGGGTCAGACTAGGAATGGGGCA........................................................................................................... | 31 | 1 | 6.00 | 6.00 | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................AGTTTGGGGACCGCCTAGTGGGCA............. | 24 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TTCTGCATCTTGTACCCAAG.................................................. | 20 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................CCCAGATCGTCGGCAGG................................................................................................................................................................................................................. | 17 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TGCAAGTGTTGTCCCAGATCGTCGGC.................................................................................................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATGGGGCAAATGTGCAGTAACGGGT.......................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTCTATCCAAGTTCTGCATCTT........................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATGGGGCAAATGTGCAGTAACG............................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATGGGGCAAATGTGCAGTAACGGTT.......................................................................................... | 25 | 1 | 2.00 | 6.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATGGGGCAAATGTGCAGTAA............................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................GGGGACCGCCTAGTGGGCAGT........... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................AGCATTGCAAGTGTTGTCCCAGATCGT........................................................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATGGGGCAAATGTGCAGTAACGGA........................................................................................... | 24 | 1 | 2.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................GGTAAGTGGGCTGCTG.................................................................................................................................................................................................. | 16 | 2 | 1.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TAGGAATGGGGCAAATGTGCAGTAACG............................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGGGGCAAATGTGCAGTAACGGGT.......................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................TGGGGCAAATGTGCAGTAACGG............................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TGCAAGTGTTGTCCCGG............................................................................................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................................TGGTGAGTTTGGGGACCGC....................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............CTAGCATTGCAAGTGTTGTCCCAGATCGTC....................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................GGGGACCGCCTAGTGGGCAGTGT......... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................GTCCCAGATCGTCGGCAGG................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TAGCATTGCAAGTGTTGTCCCAGATCGT........................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................AGCATTGCAAGTGTTGTCCCAGATCGTC....................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TAGCATTGCAAGTGTTGTCCCAGATCGTCG...................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................ATTGCAAGTGTTGTCCCAGATCGTCGGC.................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................GCATTGCAAGTGTTGTCCCAGATCGTC....................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATGGGGCAAATGTGCAGTAACGGT........................................................................................... | 24 | 1 | 1.00 | 6.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATGGGGCAAATGTGCAGTAAC.............................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TTGCAAGTGTTGTCCCAGATCGTCGG..................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TGGGGACCGCCTAGTGGGCAGT........... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TGCAAGTGTTGTCCCAGATCGTCGGCAGGGC............................................................................................................................................................................................................... | 31 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........GCACTAGCATTGCAAGT....................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................GTTGTCCCAGATCGTCGGC.................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................GCAAGTGTTGTCCCAGATCGTCGGCAG.................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................AAGTGTTGTCCCAGATCGTCG...................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................CAAGTTCTGCATCTTGTACCCAAG.................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CAAGTGTTGTCCCAGATCGTCGGCAGG................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................CCCAGATCGTCGGCAGGGCC.............................................................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................TGGGGCAAATGTGCAGTAACGGGTCA........................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................CCCAGATCGTCGGCAGGGTA.............................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................AATGGGGCAAATGTGCAGTAAC.............................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................GGTGAGTTTGGGGACCCTTG..................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .................................................................................................................................................TGGGGCAAATGTGCAGTAACGGGTCAGC...................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................CCAGATCGTCGG..................................................................................................................................................................................................................... | 12 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| ...............................................................................................................................................................................................................................................GTGGGCAGTGTCTTG..... | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCCAGCAGAGAGCACTAGCATTGCAAGTGTTGTCCCAGATCGTCGGCAGGGTAAGTGGGCTGCTGGCCTGGTCCTACCAAGCCACACCCCATCCTTCATCCCTTGATCTTAGCCTTTTCCTCCTTACTGTGGGTCAGACTAGGAATGGGGCAAATGTGCAGTAACGGGTCAGCAAAGGTTCTATCCAAGTTCTGCATCTTGTACCCAAGGCCCAGGCTGGTGAGTTTGGGGACCGCCTAGTGGGCAGTGTCTTGCGCCT .................................................................................................................................................((((((((..((((((.(((((((.(((....)))..))))..))))))))).)))).)))).................................................... ...........................................................................................................................................140..................................................................209................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | mjTestesWT4() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | mjLiverWT2() Liver Data. (liver) | SRR685340(GSM1079784) Small RNAs (15-50 nts in length) from immorta. (dicer cell line) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR553604(SRX182810) source: Spermatocytes. (Spermatocytes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR553582(SRX182788) source: Brain. (Brain) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR059765(GSM562827) DN3_control. (thymus) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | mjLiverWT3() Liver Data. (liver) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR206941(GSM723282) other. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | GSM475281(GSM475281) total RNA. (testes) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR553603(SRX182809) source: Spermatids. (Spermatids) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | GSM510452(GSM510452) newborn_rep8. (total RNA) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................GTCCCAGATCGTCGG..................................................................................................................................................................................................................... | 15 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................CCCTTGATCTTAGCCGAGT............................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .........................................................................................CATCCTTCATCCCTTTAT........................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................TATCCAAGTTCTGCAAGG............................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................GAGTTTGGGGACCGCCT..................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................GTCCCAGATCGTCGGTG................................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................CATCCCTTGATCTTAGAG................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................CTTGTACCCAAGGCCTGGA........................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................CCATCCTTCATCCCTTCA......................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................GTCCCAGATCGTCGGTAC.................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........AGCACTAGCATTGCAAGTGTTGTCCCA.............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................GATCTTAGCCTTTTCCCA......................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| .........................AGTGTTGTCCCAGATGACG....................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................GTCCCAGATCGTCGGTGAG................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................GTTGTCCCAGATCGTGGAT.................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................ATCCAAGTTCTGCATGATT.......................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................TGTCCCAGATCGTC....................................................................................................................................................................................................................... | 14 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |