| (1) AGO.mut | (11) AGO2.ip | (2) B-CELL | (13) BRAIN | (6) CELL-LINE | (2) DCR.mut | (7) EMBRYO | (9) ESC | (2) FIBROBLAST | (1) LIVER | (7) OTHER | (3) OTHER.mut | (4) PIWI.ip | (1) PIWI.mut | (2) SKIN | (4) SPLEEN | (13) TESTES | (2) THYMUS | (1) UTERUS |
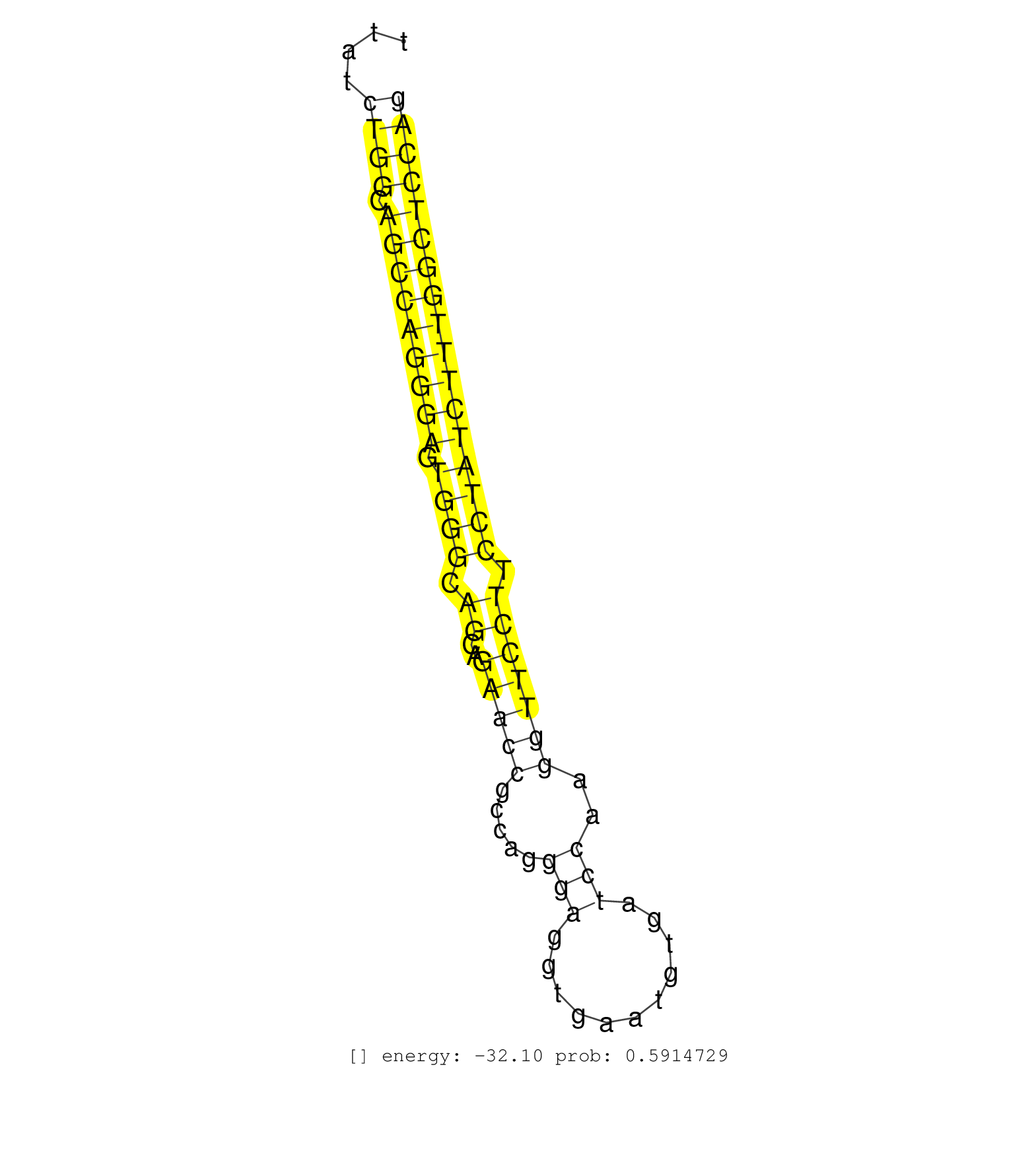
| AGCAACCTTGTCATCCACAGACGAATCCACACAGGAGAGAAACCCCTGCAGTGAGTTCCAGGGGCCATGGAGGGAGCTATTATCTGGCAGCCAGGGAGTGGGCAGCAGAACCGCCAGGGAGGTGAATGTGATCCAAGGTTCCTTCCTATCTTTGGCTCCAGGTGTGAGATATGTGGATTTACTTGCCGGCAGAAGGCCTCCCTAAACTGGC ...................................................................................((((.(((((((((.((((.((..(((((.....(((...........)))..))))))).))))))))))))))))).................................................. ...............................................................................80...............................................................................161................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR394084(GSM855970) background strain: C57BL6/SV129cell type: KRa. (dicer cell line) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR685340(GSM1079784) Small RNAs (15-50 nts in length) from immorta. (dicer cell line) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR073954(GSM629280) total RNA. (blood) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR059766(GSM562828) DN3_Drosha. (thymus) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR059768(GSM562830) Treg_control. (spleen) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | RuiDGCR8WT(Rui) DGCR8-dependent microRNA biogenesis is essent. (skin) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR206941(GSM723282) other. (brain) | SRR023849(GSM307158) NPCsmallrna_rep1. (cell line) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR553603(SRX182809) source: Spermatids. (Spermatids) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR553582(SRX182788) source: Brain. (Brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................TGGCAGCCAGGGAGTGGGCAGCAGA...................................................................................................... | 25 | 1 | 12.00 | 12.00 | - | 4.00 | - | - | - | - | - | 1.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTCCA................................................... | 22 | 1 | 11.00 | 11.00 | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTCCAGT................................................. | 24 | 1 | 9.00 | 2.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................CCTTCCTATCTTTGGCTCCAGT................................................. | 22 | 1 | 7.00 | 1.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................................................................TGAGATATGTGGATTTACTTGCCGGC..................... | 26 | 1 | 7.00 | 7.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GTTCCTTCCTATCTTTGGCTCCAG.................................................. | 24 | 1 | 5.00 | 5.00 | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CTTCCTATCTTTGGCTCCAGT................................................. | 21 | 1 | 5.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................GTGAGATATGTGGATTTACTTGCCGGC..................... | 27 | 1 | 4.00 | 4.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGGCAGCCAGGGAGTGGGCAGCAGAACCGCCAGGGAGGTGAATGTGATCCAAGGTTCCTTCCTATCTTTGGCTCCAGT................................................. | 78 | 1 | 4.00 | 1.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTCCAA.................................................. | 23 | 1 | 3.00 | 11.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................CTTCCTATCTTTGGCTCCAG.................................................. | 20 | 1 | 3.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCTTCCTATCTTTGGCTCCAGTT................................................ | 23 | 1 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CTTCCTATCTTTGGCTCCAGA................................................. | 21 | 1 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTCCAG.................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCTTCCTATCTTTGGCTCCAGA................................................. | 22 | 1 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTCC.................................................... | 21 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................AGGGAGGTGAATGTGATA.............................................................................. | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTCT.................................................... | 21 | 1 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GGGCCATGGAGGGAGGCAC................................................................................................................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ....................................................................................................................................................................TGAGATATGTGGATTTACTTGCCGGCA.................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCTTCCTATCTTTGGCTCCAGC................................................. | 22 | 1 | 2.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTCCATT................................................. | 24 | 1 | 2.00 | 11.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................TGGCAGCCAGGGAGTGGGCAGC......................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTCCTATCTTTGGCTCCA................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTCCAT.................................................. | 23 | 1 | 2.00 | 11.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCT...................................................... | 19 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCTTCCTATCTTTGGCTCCAG.................................................. | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................CAGCCAGGGAGTGGGCAGCAGAACC................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGGATTTACTTGCCGGCAGAAGGCCTC........... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGGATTTACTTGCCGGCAGAAGGCCTCC.......... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCTTCCTATCTTTGGCTCCAGTAAA.............................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGTGGATTTACTTGCCGGCAGAAGGCCT............ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCTTCCTATCTTTGGCTCCAGTAA............................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGGCAGCCAGGGAGTGGGCAGCATT...................................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................GGGAGGTGAATGTGAAACC............................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................CTTCCTATCTTTGGCTCCAGTAAC.............................................. | 24 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TCCTTCCTATCTTTGGCTCCAGA................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................CCTTCCTATCTTTGGCTCCCGT................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................TGGCAGCCAGGGAGTGGGCAGCAGAACCGCCAGGGAGGTGAATGTGATCCAAGGTTCCTTCCTATCTTTGGCTCCAG.................................................. | 77 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGGCAGCCAGGGAGTGGGCAGCAGAACCGCCAGGGAGGTGAATGTGATCCAAGGTTCCTTCCTATCTTTGGCTCCAGTTTT.............................................. | 81 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................ATGTGGATTTACTTGCCGGCAGAAGGCCTC........... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGGCAGCCAGGGAGTGGGCAGCAG....................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................AGGGAGGTGAATGTGAAACC............................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTCCAAT................................................. | 24 | 1 | 1.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGGCAGCCAGGGAGTGGGCAGCAGAACCGCCAGGGAGGTGAATGTGATCCAAGGTTCCTTCCTATCTTTGGCTCC.................................................... | 75 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TACTTGCCGGCAGAAGGCCTCCCTAAA..... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCTTCCTATCTTTGGCTCCAGTTT............................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................AGGGAGGTGAATGTGATCGAAA.......................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................CCTTCCTATCTTTGGCTCCAGTAT............................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGGCAGCCAGGGAGTGGGC............................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ACTTGCCGGCAGAAGGCCTCCCT........ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................CAGCCAGGGAGTGGGCAGCA........................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTC..................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................CAGCCAGGGAGTGGGCA........................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCTTCCTATCTTTGGCTCCAGTA................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TATGTGGATTTACTTGCCGGCAGAAGGCCTC........... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................AGATATGTGGATTTACTTGCCGGCAG................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................AGGGAGGTGAATGTGTAT.............................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................GAGATATGTGGATTTACTTGCCGGC..................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CTTCCTATCTTTGGCTCCAGTA................................................ | 22 | 1 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTCCT................................................... | 22 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTCCATA................................................. | 24 | 1 | 1.00 | 11.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TGTGGATTTACTTGCCGGCAGAAGGCCTCCCT........ | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TGTGAGATATGTGGATTTACTTGCCGGC..................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGGCAGCCAGGGAGTGGGCAGCAAT...................................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTCA.................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGGCAGCCAGGGAGTGGGCAGCAGAACCG.................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TATGTGGATTTACTTGCCGGCAGAAGGCCT............ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTCCTTCCTATCTTTGGCTCCATTT................................................ | 25 | 1 | 1.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................GTGTGAGATATGTGGAT................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGGCAGCCAGGGAGTGGG............................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................CAGGGAGGTGAATGTGAT............................................................................... | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| AGCAACCTTGTCATCCACAGACGAATCCACACAGGAGAGAAACCCCTGCAGTGAGTTCCAGGGGCCATGGAGGGAGCTATTATCTGGCAGCCAGGGAGTGGGCAGCAGAACCGCCAGGGAGGTGAATGTGATCCAAGGTTCCTTCCTATCTTTGGCTCCAGGTGTGAGATATGTGGATTTACTTGCCGGCAGAAGGCCTCCCTAAACTGGC ...................................................................................((((.(((((((((.((((.((..(((((.....(((...........)))..))))))).))))))))))))))))).................................................. ...............................................................................80...............................................................................161................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR394084(GSM855970) background strain: C57BL6/SV129cell type: KRa. (dicer cell line) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR685340(GSM1079784) Small RNAs (15-50 nts in length) from immorta. (dicer cell line) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR073954(GSM629280) total RNA. (blood) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR059766(GSM562828) DN3_Drosha. (thymus) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR059768(GSM562830) Treg_control. (spleen) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | RuiDGCR8WT(Rui) DGCR8-dependent microRNA biogenesis is essent. (skin) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR206941(GSM723282) other. (brain) | SRR023849(GSM307158) NPCsmallrna_rep1. (cell line) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR553603(SRX182809) source: Spermatids. (Spermatids) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR553582(SRX182788) source: Brain. (Brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............CACAGACGAATCCACACACG................................................................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................GCCATGGAGGGAGCTGTG.................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........TCATCCACAGACGAAT......................................................................................................................................................................................... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |