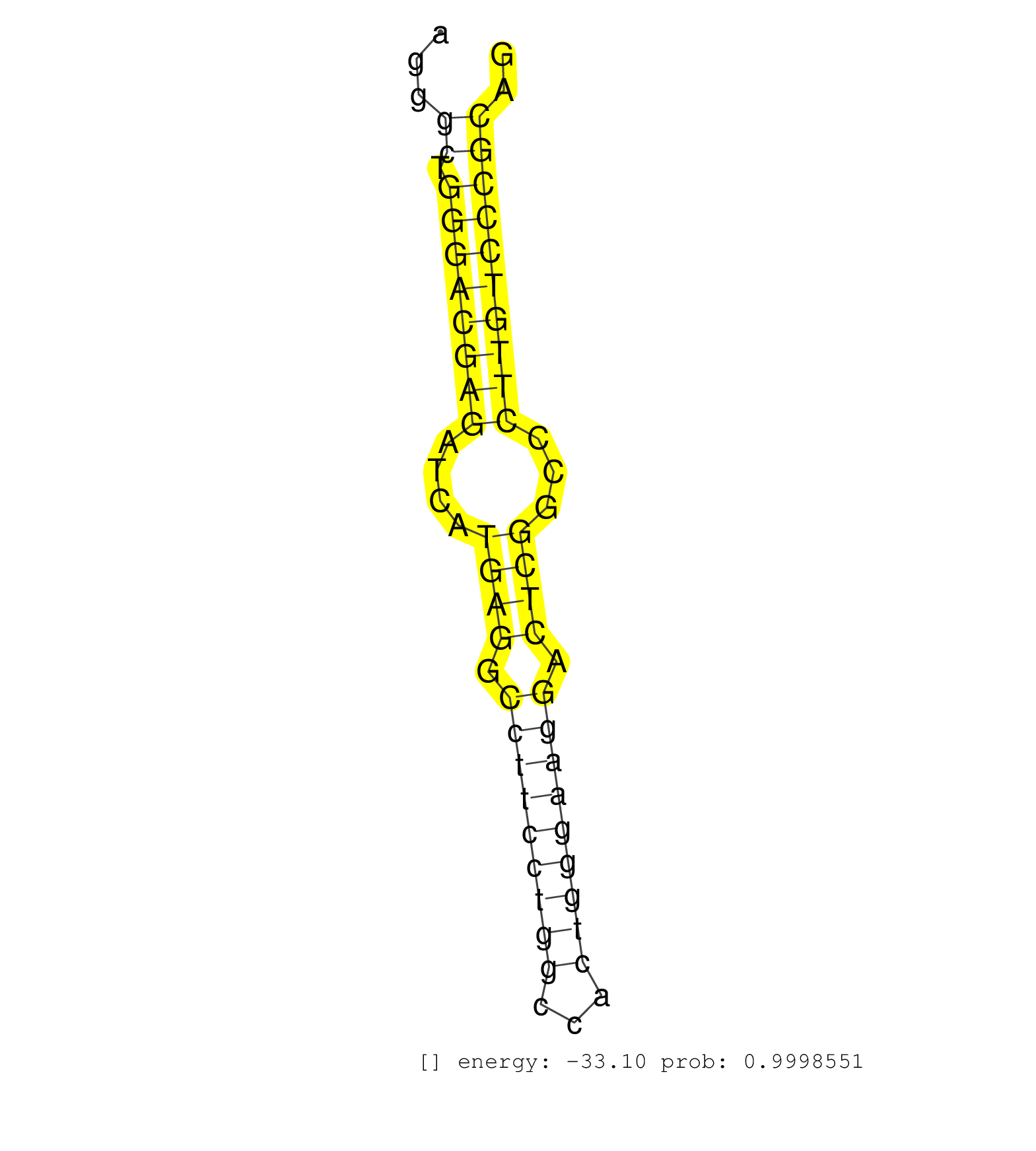
| (5) AGO2.ip | (2) B-CELL | (15) BRAIN | (7) EMBRYO | (4) ESC | (1) KIDNEY | (7) OTHER | (1) OTHER.mut | (3) TESTES | (5) TOTAL-RNA |
| CTCAAATCCTACTGCACTGAGGGATCTTAAGGAAGGAGGGCAACCAGACAGGAATGGCACTGTGCCCATGGGAGGAGCCATATCAATCACTGCTGGAATGCTCCTGGTGTGTGCAGGAGAAGCAGGTCTGGATAGTAGGGCTGGGACGAGATCATGAGGCCTTCCTGGCCACTGGGAAGGACTCGGCCCTTGTCCCGCAGGTGATGGGTGTCTTTAGTGACGAGGATGTGAAGCAGATCCTGAAGATGAT ...........................................................................................................................................((.((((((((....((((.(((((((((...))))))))).))))...)))))))))).................................................... ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | mjTestesWT4() Testes Data. (testes) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR553582(SRX182788) source: Brain. (Brain) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR206941(GSM723282) other. (brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR206939(GSM723280) other. (brain) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR206942(GSM723283) other. (brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................TGGGACGAGATCATGAGGC.......................................................................................... | 19 | 1 | 22.00 | 22.00 | 20.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGGACGAGATCATGAGGCCTTC...................................................................................... | 23 | 1 | 15.00 | 15.00 | 3.00 | 2.00 | - | - | 1.00 | 2.00 | - | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGGACGAGATCATGAGGCCTT....................................................................................... | 22 | 1 | 9.00 | 9.00 | 1.00 | 1.00 | - | 1.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................GTGACGAGGATGTGAAGCAGA............. | 21 | 1 | 8.00 | 8.00 | - | - | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGGACGAGATCATGAGGCCT........................................................................................ | 21 | 1 | 6.00 | 6.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGGACGAGATCATGAGGCC......................................................................................... | 20 | 1 | 6.00 | 6.00 | 3.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGGACGAGATCATGAGGCCTTT...................................................................................... | 23 | 1 | 5.00 | 9.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CGAGGATGTGAAGCAGGAGT.......... | 20 | 5.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | |
| ............................................................................................................................................CTGGGACGAGATCATGAGGCCTTC...................................................................................... | 24 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GGCTGGGACGAGATCATGAGGCCT........................................................................................ | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GACTCGGCCCTTGTCCCGCAG.................................................. | 21 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CTGGGACGAGATCATGAGGCCTTAA..................................................................................... | 25 | 1 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGACGAGGATGTGAAGATA.............. | 19 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................CTGGGACGAGATCATGAGGCC......................................................................................... | 21 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................AAGGACTCGGCCCTTGTCCCGC.................................................... | 22 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................AGGACTCGGCCCTTGTCCCGCAG.................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGGACGAGATCATGAGGCAT........................................................................................ | 21 | 1 | 2.00 | 22.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................AGGACTCGGCCCTTGTCCCGC.................................................... | 21 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGGACGAGATCATGAGGCCTTCT..................................................................................... | 24 | 1 | 2.00 | 15.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGCAGGAGAAGCAGGTCTGGATAGTAGGGCTGGGACGAGATCATGAGGCCTTCCTGGCCACTGGGAAGGACTCGGCCCTTGTCCCGCAGA................................................. | 90 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................CTGGGACGAGATCATGAGGCCT........................................................................................ | 22 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGGGAAGGACTCGGCGC............................................................. | 17 | 4 | 1.50 | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | 0.25 | - | - | - | - | 0.75 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................AGGACTCGGCCCTTGTCCCGTT................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................AGGACTCGGCCCTTGTCCCGCAGT................................................. | 24 | 1 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CTGGGACGAGATCATGAGGCA......................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................CTGGGACGAGATCATGAGGCAT........................................................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................AGTGACGAGGATGTGAAGC................ | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CTGGGACGAGATCATGAGGCCTTA...................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GACTCGGCCCTTGTCCCGC.................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GGCTGGGACGAGATCATGAGGGGT........................................................................................ | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................GACTCGGCCCTTGTCCCGCAGT................................................. | 22 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGGACGAGATCATGAGGCCTTCA..................................................................................... | 24 | 1 | 1.00 | 15.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGCAGGAGAAGCAGGTCTGGTT..................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................CTGGGACGAGATCATGAGGCCTT....................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CTGGGACGAGATCATGAGGCCTTCATT................................................................................... | 27 | 1 | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................GACGAGGATGTGAAGATTT............. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................TGAAGCAGATCCTGAAGATGA. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................AGCAGGTCTGGATAGGGC................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................AAGGACTCGGCCCTTGTCCCGCAGT................................................. | 25 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TGGGACGAGATCATGAGGCA......................................................................................... | 20 | 1 | 1.00 | 22.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGGACGAGATCATGAGGCCTTA...................................................................................... | 23 | 1 | 1.00 | 9.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CGAGGATGTGAAGCAGGAG........... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TGGGACGAGATCATGAGGCCTAT...................................................................................... | 23 | 1 | 1.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGACGAGGATGTGAAT................. | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................GCTGGGACGAGATCATGAGGC.......................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGGACGAGATCATGAGGCCTTAT..................................................................................... | 24 | 1 | 1.00 | 9.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GACTCGGCCCTTGTCCCGCAGA................................................. | 22 | 1 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CGAGGATGTGAAGCAGGAGC.......... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ........................................................................................................................................................................................................................GTGACGAGGATGTGAAGCAGATC........... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................AAGGACTCGGCCCTTGTCCCGT.................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TGGGACGAGATCATGAGGCCTTCAT.................................................................................... | 25 | 1 | 1.00 | 15.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................TGGGACGAGATCATGAGGCCTAA...................................................................................... | 23 | 1 | 1.00 | 6.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGGGAAGGACTCGGCG.............................................................. | 16 | 4 | 1.00 | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |
| ................................................................................................................................................................................AAGGACTCGGCCCTTGTCCCGCT................................................... | 23 | 1 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGGACGAGATCATGAGGCCTTTA..................................................................................... | 24 | 1 | 1.00 | 9.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................AAGGACTCGGCCCTTGTCCCGCAT.................................................. | 24 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................GTCTTTAGTGACGAGGAT....................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGGGAAGGACTCGGC............................................................... | 15 | 4 | 0.75 | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGGGAAGGACTCGGCGCTC........................................................... | 19 | 4 | 0.75 | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| ............................................................................................................................................................................TGGGAAGGACTCGGCT.............................................................. | 16 | 4 | 0.50 | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGGGAAGGACTCGGCGCT............................................................ | 18 | 4 | 0.25 | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGGGAAGGACTCGGCGCCC........................................................... | 19 | 4 | 0.25 | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGGGAAGGACTCGGCACT............................................................ | 18 | 4 | 0.25 | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................GGAAGGACTCGGCC.............................................................. | 14 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - |
| CTCAAATCCTACTGCACTGAGGGATCTTAAGGAAGGAGGGCAACCAGACAGGAATGGCACTGTGCCCATGGGAGGAGCCATATCAATCACTGCTGGAATGCTCCTGGTGTGTGCAGGAGAAGCAGGTCTGGATAGTAGGGCTGGGACGAGATCATGAGGCCTTCCTGGCCACTGGGAAGGACTCGGCCCTTGTCCCGCAGGTGATGGGTGTCTTTAGTGACGAGGATGTGAAGCAGATCCTGAAGATGAT ...........................................................................................................................................((.((((((((....((((.(((((((((...))))))))).))))...)))))))))).................................................... ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | mjTestesWT4() Testes Data. (testes) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR553582(SRX182788) source: Brain. (Brain) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR206941(GSM723282) other. (brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR206939(GSM723280) other. (brain) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR206942(GSM723283) other. (brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................................ACGAGGATGTGAAGCG............... | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................GGCCCTTGTCCCGCAAAC................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |