| (1) HEART | (2) KIDNEY | (3) OTHER | (1) TESTES |
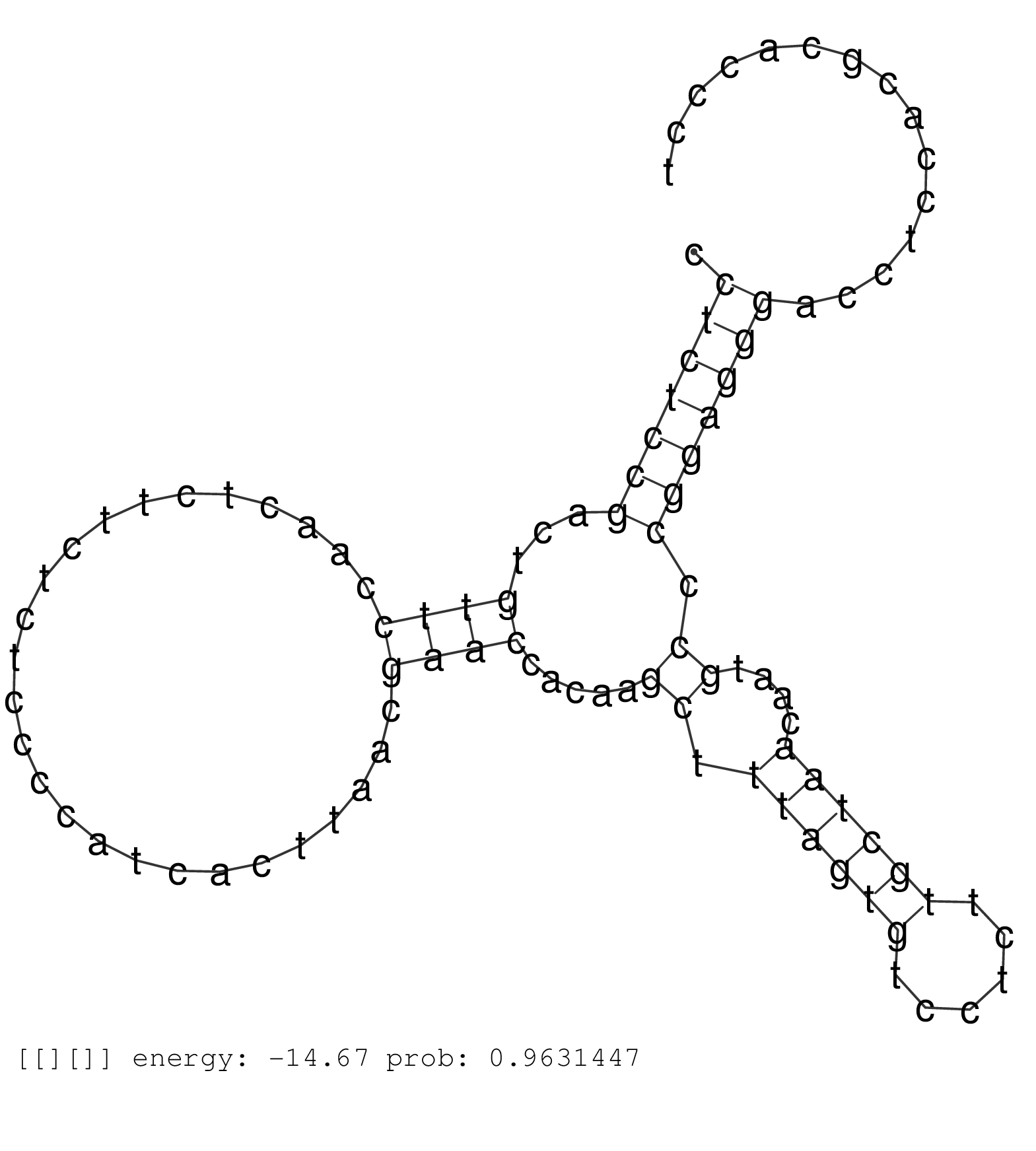
| ATCGACTTGGAGTGGGTGGTGAGCGCCACCACCACGGGCACCATTTCCCAGTGAGTCCCCCCTGTGGCCCCGCCTCACTGCTCCGCCCCCTGCAGTGACACCCTGTCCCCCAGCCCTGCCGTGGTCCTGCTGTGCCTCCACTGGGGTGCTTCCCCTCCCCCAGCTTCCTCCGGCACACACAAGCCCCGTTCCTGTGGCTGCCTCTCCGACTGTTCCAACTCTTCTCTCCCCATCACTTAACGAACCACAAGCTTTAGTGTCCTCTTGCTAACAATGCCCGGAGGGACCTCCACGCACCCTGGAGACTCTTCGGGAAGGGGGCCGCCACTAGAAAGGGGCGGCACATACGA .........................................................................................................................................................................................................(((((((...((((..........................)))).....((.((((((......))))))....)).)))))))................................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553579(SRX182785) source: Heart. (Heart) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553581(SRX182787) source: Testis. (testes) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................CCCCCTGCAGTGACACCA....................................................................................................................................................................................................................................................... | 18 | 11.00 | 1.00 | 7.00 | 1.00 | 2.00 | - | 1.00 | - | - | |
| ......................................................................................CCCCTGCAGTGACACCA....................................................................................................................................................................................................................................................... | 17 | 10.00 | 1.00 | 5.33 | 2.00 | 0.67 | 1.33 | 0.33 | - | 0.33 | |
| ......................................................................................CCCCTGCAGTGACACC........................................................................................................................................................................................................................................................ | 16 | 3 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - |
| ..........................................................................................TGCAGTGACACCCTGAAC.................................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | |
| .....................................................................................CCCCCTGCAGTGACACC........................................................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - |
| ......................................................................................CCCCTGCAGTGACACCAA...................................................................................................................................................................................................................................................... | 18 | 0.33 | 1.00 | 0.33 | - | - | - | - | - | - | |
| .....................................................................................CCCCCTGCAGTGACA.......................................................................................................................................................................................................................................................... | 15 | 10 | 0.30 | 0.30 | 0.10 | - | - | 0.20 | - | - | - |
| ......................................................................................CCCCTGCAGTGACAC......................................................................................................................................................................................................................................................... | 15 | 7 | 0.14 | 0.14 | - | - | 0.14 | - | - | - | - |
| ATCGACTTGGAGTGGGTGGTGAGCGCCACCACCACGGGCACCATTTCCCAGTGAGTCCCCCCTGTGGCCCCGCCTCACTGCTCCGCCCCCTGCAGTGACACCCTGTCCCCCAGCCCTGCCGTGGTCCTGCTGTGCCTCCACTGGGGTGCTTCCCCTCCCCCAGCTTCCTCCGGCACACACAAGCCCCGTTCCTGTGGCTGCCTCTCCGACTGTTCCAACTCTTCTCTCCCCATCACTTAACGAACCACAAGCTTTAGTGTCCTCTTGCTAACAATGCCCGGAGGGACCTCCACGCACCCTGGAGACTCTTCGGGAAGGGGGCCGCCACTAGAAAGGGGCGGCACATACGA .........................................................................................................................................................................................................(((((((...((((..........................)))).....((.((((((......))))))....)).)))))))................................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553579(SRX182785) source: Heart. (Heart) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553581(SRX182787) source: Testis. (testes) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................TGGAACAGTCGGAGAGGCAGCCACAGG..................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - |