| (1) KIDNEY | (1) OTHER | (2) TESTES |
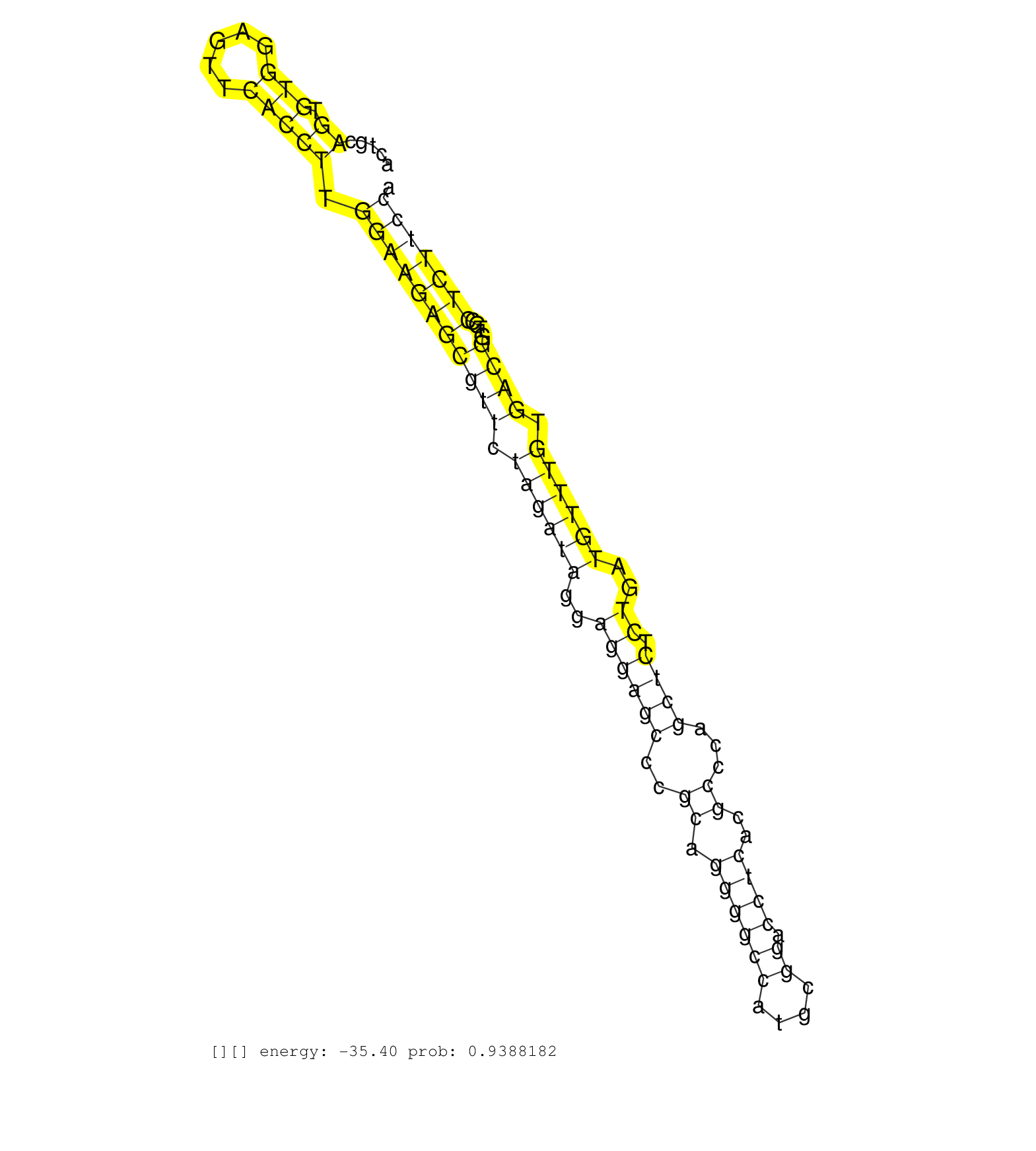
| GGCCCCCTCCCCCCAGGCTCCTTTTTTGAGTGACAGCTTTCTGGGCTGTCACGGTCCAATCAGAAACACCCCTCTGTCTAGGACTGCAGTGTGGAGTTCACCTTGGAAGAGCGTTCTAGATAGGAGGAGCCCGCAGGGGCCATGCGGACCTCACGCCCAGCTCTCTGATGTTTGTGACGGTGCCTCTTCCAATGAACATTCCCAGCCCAGCCCCAGCACTCCAACAGACCCTGCCCCTCGTGAGCTGGATAGACTTGGGGTGGGGAGGGAGGGAGTTTGTCTGTCTCCCTCCCCTCTCAGAACGTACTGATTGGAAGGCGCGTGTTCAGCAGAACCTGCACACAGGACAG .......................................................................................((.(((.....))))).(((((((((((.((((((..((((((..((.((((((....)).))))..))...)))).))..)))))).))))....)))))))................................................................................................................................................................ ..................................................................................83..........................................................................................................191............................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553607(SRX182813) source: Testis. (testes) |
|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................................................................................................TCTCAGAACGTACTGATTGGAAGGC............................... | 25 | 1 | 8.00 | 8.00 | 8.00 | - | - | - |
| ......................................................................................................................................................................................................................................................................................................TCTCAGAACGTACTGATTGGAAGGCGC............................. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - |
| ...................................................................................................................................................................TCTGATGTTTGTGACGGTGCCTCTTC................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .......................................................................................AGTGTGGAGTTCACCTTGGAAGAGC.............................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..................................................................................................................................................................CTCTGATGTTTGTGACGGTGCCTCT................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...................................................................................................................................................................TCTGATGTTTGTGACGGTGCCTCTT.................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................AGAACGTACTGATTGGAAGGCGCGTG.......................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .......................................................................................................................................................................................................................................................................................................CTCAGAACGTACTGATTGGAAGGCGC............................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ......................................................................................................................................................................................................................................................................................................TCTCAGAACGTACTGATTGGAAGGCA.............................. | 26 | 1.00 | 8.00 | 1.00 | - | - | - | |
| ...................................................................................................................................................................TCTGATGTTTGTGACGGT......................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................GAAGGCGCGTGTTCAGCAGAACCTGC........... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................CGTACTGATTGGAAGG................................ | 16 | 2 | 0.50 | 0.50 | 0.50 | - | - | - |
| GGCCCCCTCCCCCCAGGCTCCTTTTTTGAGTGACAGCTTTCTGGGCTGTCACGGTCCAATCAGAAACACCCCTCTGTCTAGGACTGCAGTGTGGAGTTCACCTTGGAAGAGCGTTCTAGATAGGAGGAGCCCGCAGGGGCCATGCGGACCTCACGCCCAGCTCTCTGATGTTTGTGACGGTGCCTCTTCCAATGAACATTCCCAGCCCAGCCCCAGCACTCCAACAGACCCTGCCCCTCGTGAGCTGGATAGACTTGGGGTGGGGAGGGAGGGAGTTTGTCTGTCTCCCTCCCCTCTCAGAACGTACTGATTGGAAGGCGCGTGTTCAGCAGAACCTGCACACAGGACAG .......................................................................................((.(((.....))))).(((((((((((.((((((..((((((..((.((((((....)).))))..))...)))).))..)))))).))))....)))))))................................................................................................................................................................ ..................................................................................83..........................................................................................................191............................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553607(SRX182813) source: Testis. (testes) |
|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................gaGAGAGCTGGGCGTGAG....................................................................................................................................................................................... | 18 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ...........tcaAAAAAGGAGCCTACT................................................................................................................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ..........AAAAGGAGCCTGGGG..................................................................................................................................................................................................................................................................................................................................... | 15 | 16 | 0.06 | 0.06 | 0.06 | - | - | - |