| (1) HEART | (2) KIDNEY | (1) OTHER | (2) TESTES |
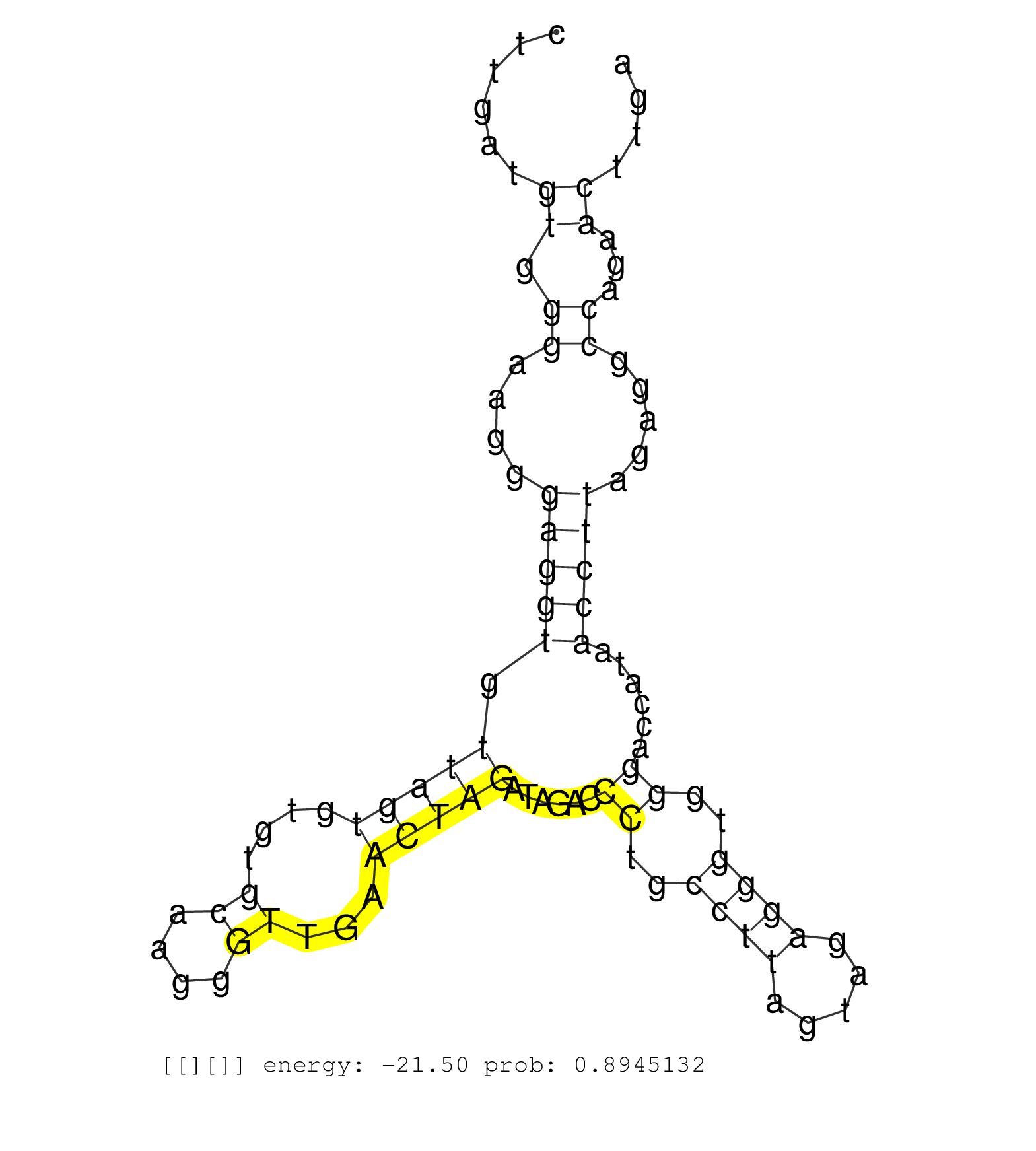
| CGTGAGAGCTATCTGGGGGAAAGAATCCTTACCAAGGTTTTTTTGGAAAGGTACGAATCTTACCTTTTTTCCCCTTCTGTGTCTCAGGGTAATACTCTATTCAGAGTCGCCCCTTTGCTCATTTTCTCCCATATTTGTTACCTTCCTGAGGCCTCAGTATTAGTCATGAGCACAAAGTTTGGAGACCCTTGGCGTTGTTTCTTGATGTGGGAAGGGAGGTGTTAGTGTGTGCAAGGGTTGAACTAGATAGACCCTGCCTTAGTAGAGGGTGGGACCATAACCTTAGAGGCCAGAACTTGATCCAGAAGTTGCTGTCCACAGAAGTGCTTTCTAGTTCATCATTTTTGTCT ..............................................................................................................................................................................................................((.((....(((((.(((((....((....))...)))))......((..((((.....))))..))......))))).....))...))...................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553607(SRX182813) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553580(SRX182786) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................................GTTGAACTAGATAGACCC................................................................................................ | 18 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - |
| ..............................................................................................................................................................................................................................................TGAACTAGATAGACCCTGCC............................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................................................................................................TGTTAGTGTGTGCAAGGGTTGAACTAGATA..................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................................TGAGCACAAAGTTTGGAGACCCTTGGC............................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................TGATCCAGAAGTTGCTGTCCACAGAAGTGC....................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................CAGAACTTGATCCAGAGCCC........................................ | 20 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | |
| ..........................................................................................................................................................................................................................................................................................TTAGAGGCCAGAACTTGATCCAGAAGTTGC...................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................................................TGAACTAGATAGACCCTG.............................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................TATTAGTCATGAGCACAAAGTTTGGAGACC................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................................................................................................GTAGAGGGTGGGACCA......................................................................... | 16 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................................................................................................TAGTAGAGGGTGGGACCATAACCTTAGAGGC............................................................ | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................TGAACTAGATAGACCCTGCCTTAGTAGAGGGT................................................................................ | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................TGAACTAGATAGACCCTGCCTTA......................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................................................................................ACTAGATAGACCCTGCCTTAGTAGAGGGT................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................................................................................................TAGATAGACCCTGCCTTAGTAGAGGGTGGT............................................................................. | 30 | 1.00 | 0.00 | 1.00 | - | - | - | - | - |
| CGTGAGAGCTATCTGGGGGAAAGAATCCTTACCAAGGTTTTTTTGGAAAGGTACGAATCTTACCTTTTTTCCCCTTCTGTGTCTCAGGGTAATACTCTATTCAGAGTCGCCCCTTTGCTCATTTTCTCCCATATTTGTTACCTTCCTGAGGCCTCAGTATTAGTCATGAGCACAAAGTTTGGAGACCCTTGGCGTTGTTTCTTGATGTGGGAAGGGAGGTGTTAGTGTGTGCAAGGGTTGAACTAGATAGACCCTGCCTTAGTAGAGGGTGGGACCATAACCTTAGAGGCCAGAACTTGATCCAGAAGTTGCTGTCCACAGAAGTGCTTTCTAGTTCATCATTTTTGTCT ..............................................................................................................................................................................................................((.((....(((((.(((((....((....))...)))))......((..((((.....))))..))......))))).....))...))...................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553607(SRX182813) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553580(SRX182786) source: Kidney. (Kidney) |
|---|