| (1) HEART | (1) KIDNEY | (3) OTHER | (1) TESTES |
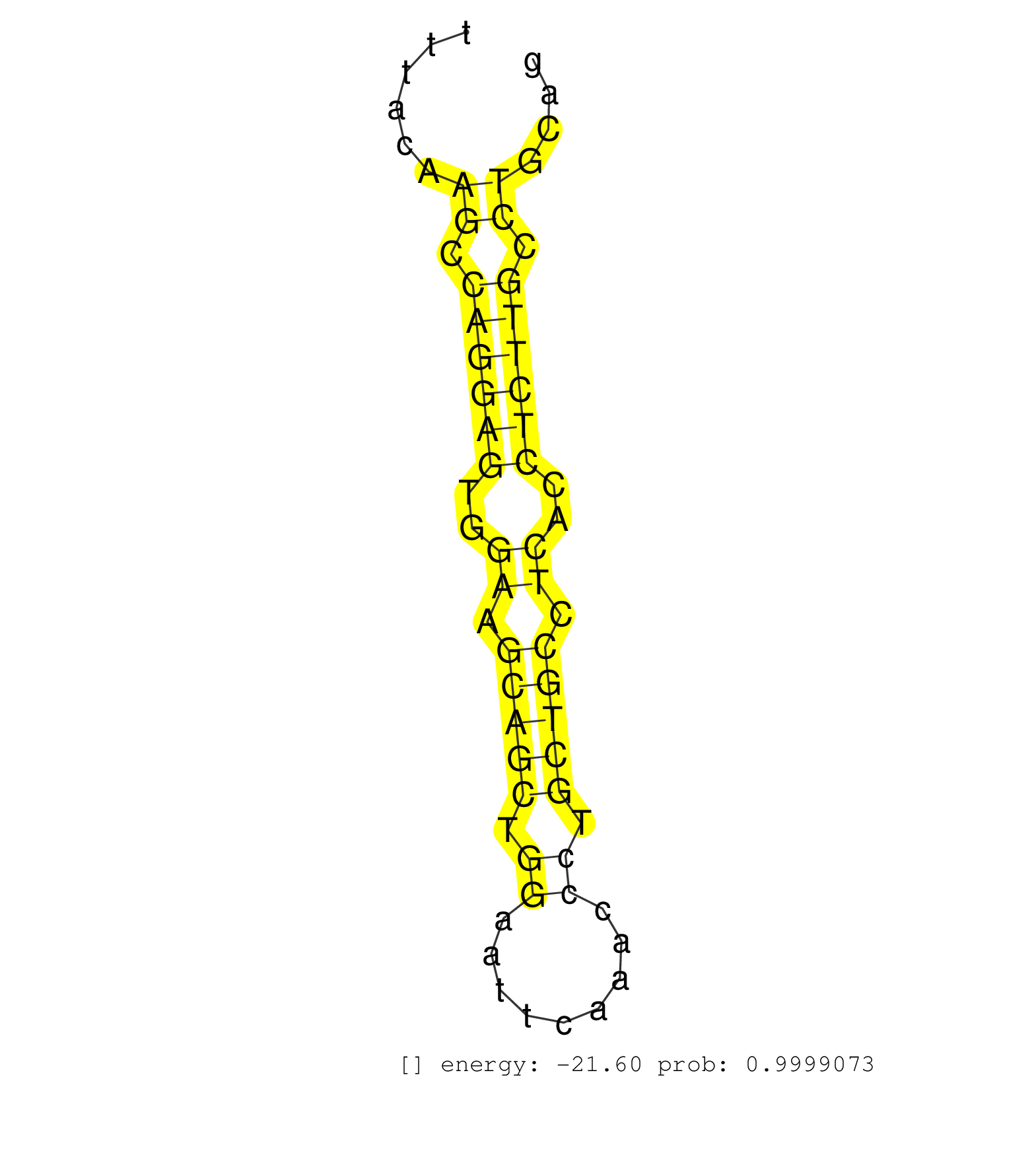
| GGCCAGAGACTGTGGTAGAACCTAAGCCTGGAGACAGTAAGAGGGTGGGGGCTTATCCATGTAGCTAGTGTGATCAGGTGGGCTTCCTAGGAAAAGGTGCCCAAGGGAGCTGTGCAGGATGGGTAGGTTTTACATCCCAGAGGCAGATATGAGAGGGTGTTCCAGGCTAATGATGAGCAGAGGCTTGGAAATAGGGTTGATCGGGTCTGGAGGGTGGAGAGGAGCCTGGGGTGAGGCTTTACAAGCCAGGAGTGGAAGCAGCTGGAATTCAAACCCTGCTGCCTCACCTCTTGCCTGCAGACTTTGGTTTCGCACAGCACATGTCCCCGTGGGATGAGAAGCACGTGCTC ...................................................................................................................................................................................................................................................((.((((((..((.(((((.((.........)).))))).))..)))))).))...................................................... .............................................................................................................................................................................................................................................238...........................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553581(SRX182787) source: Testis. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................................................AAGCCAGGAGTGGAAGCAGCTGA..................................................................................... | 23 | 12.00 | 5.00 | 4.00 | 3.00 | 3.00 | - | 1.00 | 1.00 | |
| ..................................................................................................................................................................................................................................................AAGCCAGGAGTGGAAGCAGCTG...................................................................................... | 22 | 1 | 5.00 | 5.00 | 3.00 | 2.00 | - | - | - | - |
| ...............................................................................................................................................................................................................................................TACAAGCCAGGAGTGGAAGCAGC........................................................................................ | 23 | 1 | 4.00 | 4.00 | 1.00 | 3.00 | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................TGCTGCCTCACCTCTTGCCTGC.................................................... | 22 | 1 | 3.00 | 3.00 | 1.00 | - | - | 2.00 | - | - |
| ...............................................................................................................................................................................................................................................TACAAGCCAGGAGTGGAAGCAG......................................................................................... | 22 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | 1.00 | - | - |
| ................................................................................................................................................................................................................................................ACAAGCCAGGAGTGGAAGCAGA........................................................................................ | 22 | 2.00 | 1.00 | 2.00 | - | - | - | - | - | |
| ................................................................................................................................................................................................................................................ACAAGCCAGGAGTGGAAGCAGC........................................................................................ | 22 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................TGCTGCCTCACCTCTTGCCTGCAG.................................................. | 24 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - |
| ..................................................................................................................................................................................................................................................AAGCCAGGAGTGGAAGCAGCTGT..................................................................................... | 23 | 2.00 | 5.00 | 1.00 | 1.00 | - | - | - | - | |
| ................................................................................................................................................................................................................................................ACAAGCCAGGAGTGGAAGA........................................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | |
| ..................................................................................................................................................................................................................................................AAGCCAGGAGTGGAAGCAGCT....................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................GCCAGGAGTGGAAGCAGCTGG..................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................................................................................AGCCAGGAGTGGAAGCAGCTGA..................................................................................... | 22 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | |
| ....................................................................................................................................................................................................................................................................................TGCTGCCTCACCTCTTGCCTGCA................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - |
| ................................................................................................................................................................................................................................................ACAAGCCAGGAGTGGAAGCAG......................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................TACAAGCCAGGAGTGGAAGCAGA........................................................................................ | 23 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | |
| ..................................................................................................................................................................................................................................................AAGCCAGGAGTGGAAGCAGCTGG..................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................................................................................TACAAGCCAGGAGTGGAAGCATA........................................................................................ | 23 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | |
| ..................................................................................................................................................................................................................................................AAGCCAGGAGTGGAAGCAGCTTA..................................................................................... | 23 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | |
| ....................................................................................................................................................................................................................................................................................TGCTGCCTCACCTCTTGCCTGCAGAT................................................ | 26 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | |
| ..................................................................................................................................................................................................................................................AAGCCAGGAGTGGAAGCAGCTGAA.................................................................................... | 24 | 1.00 | 5.00 | - | - | 1.00 | - | - | - |
| GGCCAGAGACTGTGGTAGAACCTAAGCCTGGAGACAGTAAGAGGGTGGGGGCTTATCCATGTAGCTAGTGTGATCAGGTGGGCTTCCTAGGAAAAGGTGCCCAAGGGAGCTGTGCAGGATGGGTAGGTTTTACATCCCAGAGGCAGATATGAGAGGGTGTTCCAGGCTAATGATGAGCAGAGGCTTGGAAATAGGGTTGATCGGGTCTGGAGGGTGGAGAGGAGCCTGGGGTGAGGCTTTACAAGCCAGGAGTGGAAGCAGCTGGAATTCAAACCCTGCTGCCTCACCTCTTGCCTGCAGACTTTGGTTTCGCACAGCACATGTCCCCGTGGGATGAGAAGCACGTGCTC ...................................................................................................................................................................................................................................................((.((((((..((.(((((.((.........)).))))).))..)))))).))...................................................... .............................................................................................................................................................................................................................................238...........................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553581(SRX182787) source: Testis. (testes) |
|---|