| (1) HEART | (2) KIDNEY | (3) OTHER | (2) TESTES |
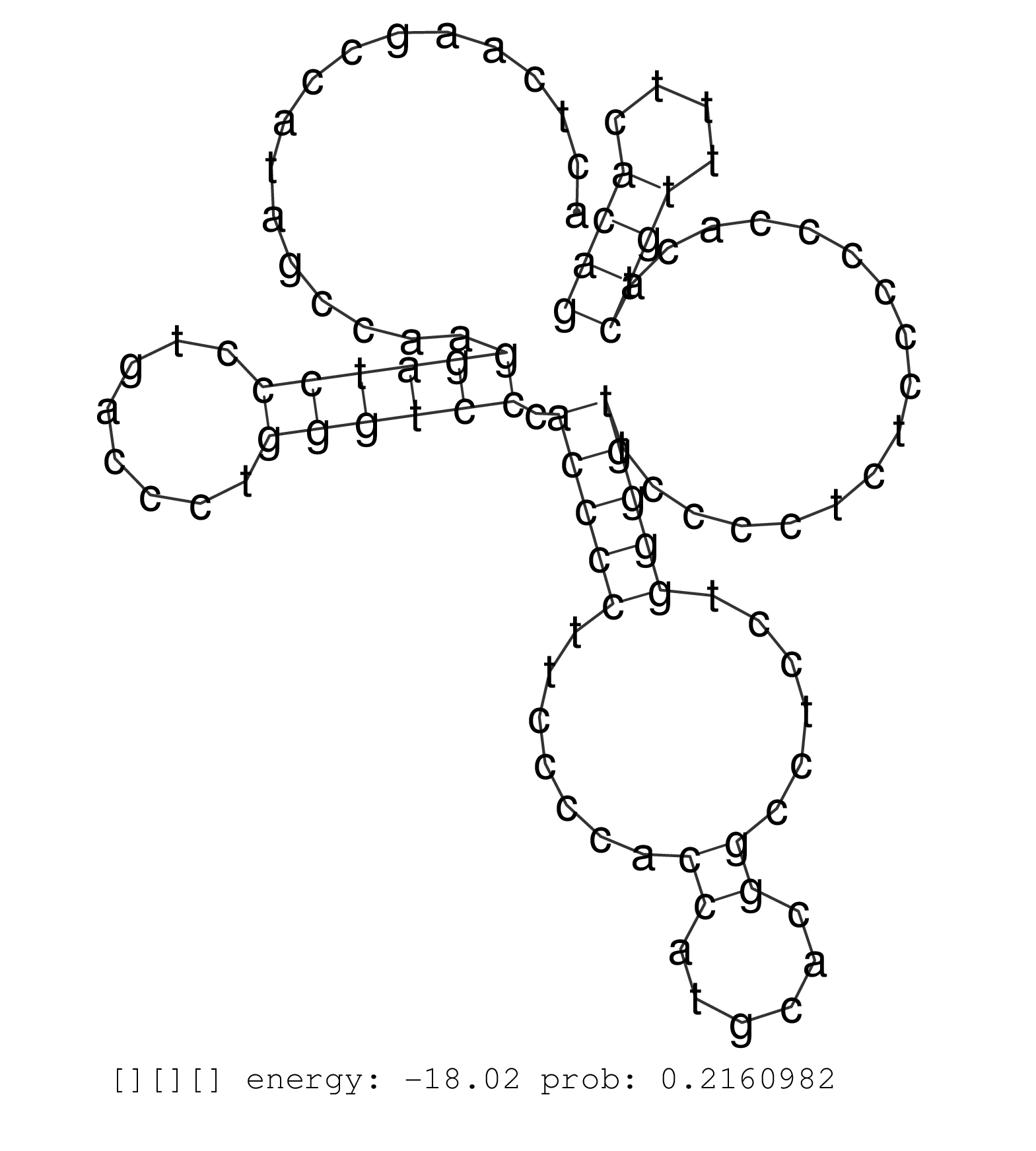
| GGCAGACACCCATGAAGGATGGTTTGGGGGAAGACCCGAGGAATGCCAGGCTGTGAGCCTCTCTCTGAGATCAGAACGGGGTCACTGATCCTCAGGGTCACTGTGTCAGTACCAGGAGGGCCTGAGAGATGGTATAGACTGGAGAATGAGGGTCAGAGAGCGAGAGTGCCCCCACCTCAGGTCATGAAGCTCACCCAGAAACTCAAGCCATAGCCAAGGATCCCTGACCCTGGGTCCCACCCCTTCCCCACCATGCACGGCCTCCTGGGGTTCCCCTCTCCCCCCACACTGTTTTCACAGTGATCAAAGCATCACGTCCTGTGACACCAGAGCCATTGACGCCATATCCC .........................................................................................................................................................................................................................((((((........)))))).(((((.......((......))......))))).................((((....)))).................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGACTTT..................................................................................................................................................................................... | 23 | 119.00 | 2.00 | 46.00 | 23.00 | 20.00 | 11.00 | 6.00 | 9.00 | 2.00 | 2.00 | |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGACTT...................................................................................................................................................................................... | 22 | 10.00 | 2.00 | 3.00 | 4.00 | - | 1.00 | 2.00 | - | - | - | |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGACT....................................................................................................................................................................................... | 21 | 6.00 | 2.00 | 1.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGACA....................................................................................................................................................................................... | 21 | 6.00 | 2.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGACTA...................................................................................................................................................................................... | 22 | 4.00 | 2.00 | 1.00 | 1.00 | - | - | 2.00 | - | - | - | |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGAC........................................................................................................................................................................................ | 20 | 3.00 | 2.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGACTTA..................................................................................................................................................................................... | 23 | 3.00 | 2.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGAAA....................................................................................................................................................................................... | 21 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGA......................................................................................................................................................................................... | 19 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGAA........................................................................................................................................................................................ | 20 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGATT....................................................................................................................................................................................... | 21 | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGAAAA...................................................................................................................................................................................... | 22 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGACTGT..................................................................................................................................................................................... | 23 | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGA........................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAG.......................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................AGGGTCAGAGAGCGAGACT....................................................................................................................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | |
| ..................................................................................................................................................TGAGGGTCAGAGAGCGAGAAAAA..................................................................................................................................................................................... | 23 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - |
| GGCAGACACCCATGAAGGATGGTTTGGGGGAAGACCCGAGGAATGCCAGGCTGTGAGCCTCTCTCTGAGATCAGAACGGGGTCACTGATCCTCAGGGTCACTGTGTCAGTACCAGGAGGGCCTGAGAGATGGTATAGACTGGAGAATGAGGGTCAGAGAGCGAGAGTGCCCCCACCTCAGGTCATGAAGCTCACCCAGAAACTCAAGCCATAGCCAAGGATCCCTGACCCTGGGTCCCACCCCTTCCCCACCATGCACGGCCTCCTGGGGTTCCCCTCTCCCCCCACACTGTTTTCACAGTGATCAAAGCATCACGTCCTGTGACACCAGAGCCATTGACGCCATATCCC .........................................................................................................................................................................................................................((((((........)))))).(((((.......((......))......))))).................((((....)))).................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553578(SRX182784) source: Cerebellum. (Cerebellum) | SRR553577(SRX182783) source: Frontal Cortex. (Frontal Cortex) | SRR553605(SRX182811) source: Frontal Cortex. (Frontal Cortex) | SRR553580(SRX182786) source: Kidney. (Kidney) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553579(SRX182785) source: Heart. (Heart) | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) |
|---|