| (1) TESTES |
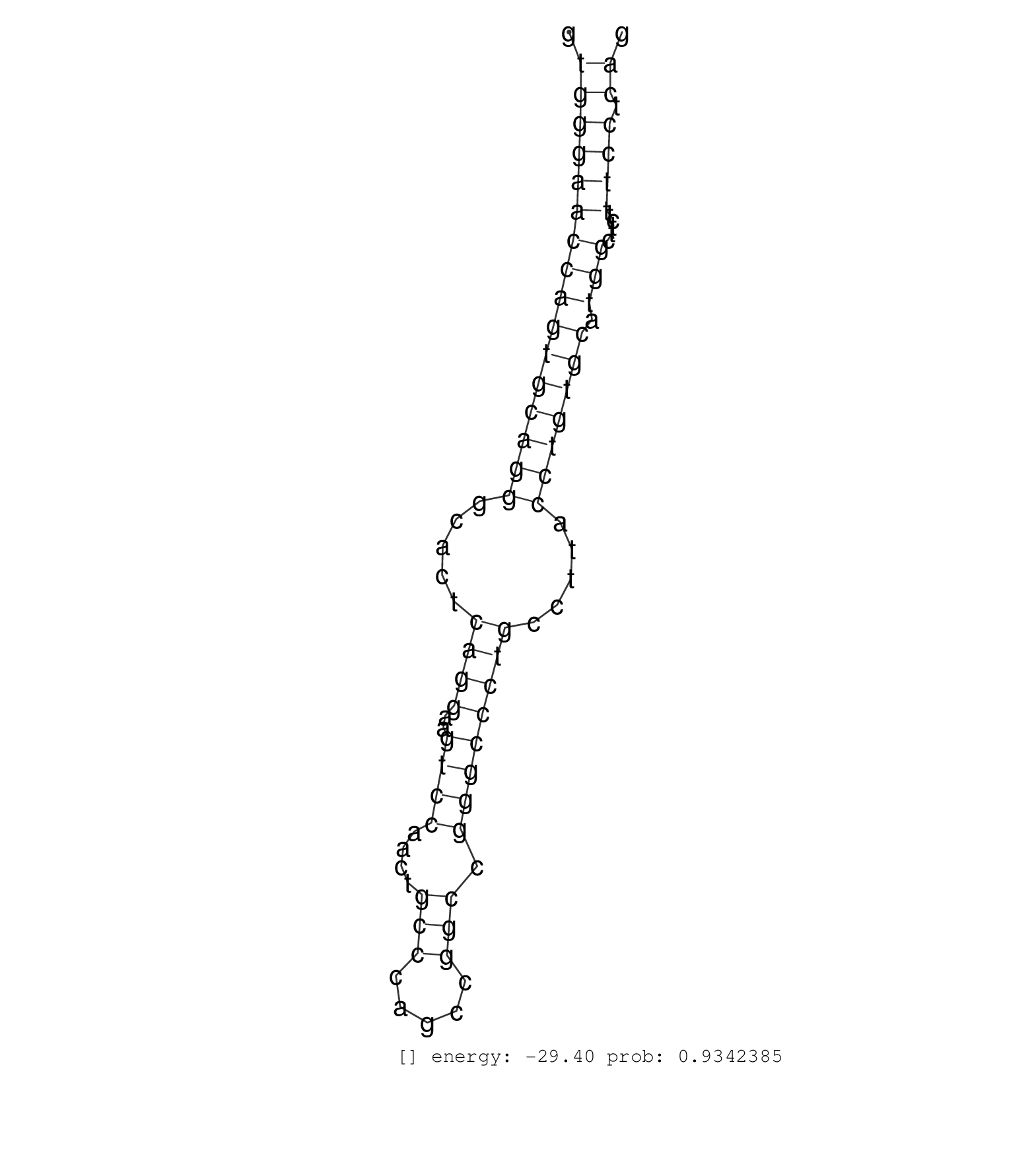
| CCTGGAGCCCCAGCAGTCAGTTCCTGGCGGTTGGGAGCTACGATGGAAAGGTGGGAACCAGTGCAGGGCACTCAGGAAGTCCAACTGCCCAGCCGGCCGGGCCCTGCCTTACCTGTGCATGGCTTCTTTCCTCAGGTGCGCATCCTTAATCACGTGACTTGGAAAATGATCACGGAGTTTGGGCA ...................................................((((((((((((((((.....((((..((((....(((.....))).)))))))).....))))))).))).....)))).))................................................... ..................................................51..................................................................................135................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) |
|---|---|---|---|---|---|
| ..............................................................................................................................................................TTGGAAAATGATCACGGAGTTTGGGC. | 26 | 1 | 1.00 | 1.00 | 1.00 |
| .................................................................................................................................................................GAAAATGATCACGGAGTTTGGCATC | 25 | 1.00 | 0.00 | 1.00 | |
| ..............................................................................................................................................TCCTTAATCACGTGACTTGGAAAATGATC.............. | 29 | 1 | 1.00 | 1.00 | 1.00 |
| CCTGGAGCCCCAGCAGTCAGTTCCTGGCGGTTGGGAGCTACGATGGAAAGGTGGGAACCAGTGCAGGGCACTCAGGAAGTCCAACTGCCCAGCCGGCCGGGCCCTGCCTTACCTGTGCATGGCTTCTTTCCTCAGGTGCGCATCCTTAATCACGTGACTTGGAAAATGATCACGGAGTTTGGGCA ...................................................((((((((((((((((.....((((..((((....(((.....))).)))))))).....))))))).))).....)))).))................................................... ..................................................51..................................................................................135................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) |
|---|---|---|---|---|---|
| ...................................................................................................................................ACGTGATTAAGGATGCGCACCTGA.............................. | 24 | 1 | 1.00 | 1.00 | 1.00 |
| .........................................................................................................................................TCCAAGTCACGTGATTAAGGATGCGC...................... | 26 | 1 | 1.00 | 1.00 | 1.00 |