| (1) HEART | (1) KIDNEY | (2) TESTES |
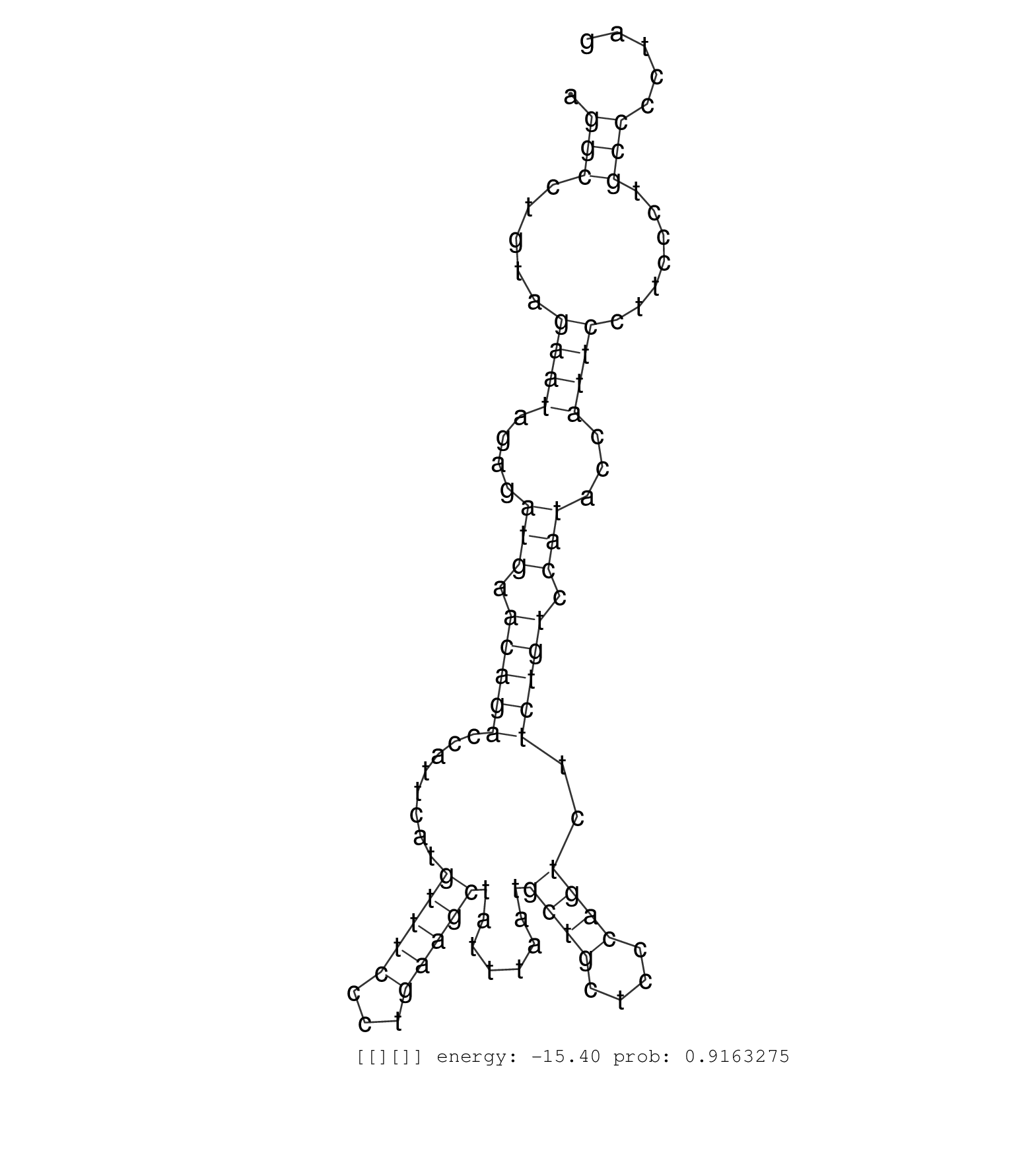
| GTTTCTTTTAGTCTTCAGATATTTTGTTCTGGTCGGTCCCTAATCTATAGTTTTGAAACTCGTGCCGCACTTGAAAGGGAAACATCCTACTACCTTTCTTTGCTCTCCATTTCCCACAAACCCTTTCCTTCATCGTCACTCCCTCCATTCTTCTCAGCATATAGATACAAGACCCTTCTTTCTATTATTTGTTGAGAGCTAGGCCTGTAGAATAGAGATGAACAGACCATTCATGTTTCCCTGAAGCTATTTAATGCTGCTCCCAGTCTTCTGTCCATACCATTCCTTCCCTGCCCCTAGGAGTGGGGTTAAGGTCTGTGACATGCAGACTGTAAGCACAGTCAGAATCT .........................................................................................................................................................................................................(((.....((((....(((.(((((........(((((...)))))........((((....))))..))))).)))...)))).......)))....................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553579(SRX182785) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................TGTAGAATAGAGATGAACAGACCATTCATGT.................................................................................................................. | 31 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - |
| ......................................................................................................................................................................................TATTATTTGTTGAGAGCTAGGCCTGTAGA........................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 2.00 | - | - |
| ......................................................................................................................................................................................TATTATTTGTTGAGAGCTAGGCCTGT.............................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - |
| ...............................................................................................................................................................................................................TAGAATAGAGATGAACAGACCATTCATGT.................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ................................................................................................................................................................................................TGAGAGCTAGGCCTGTAGAATAGAGATGAACT.............................................................................................................................. | 32 | 1.00 | 1.00 | 1.00 | - | - | - | |
| ....................................................................................................................................................................................................AGCTAGGCCTGTAGAATAGAGATGAACAG............................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| .......................................................................................................................................................................................................TAGGCCTGTAGAATAGAGATGAACAGACC.......................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .............................................................................................................................................................................................................TGTAGAATAGAGATGAACAGACCATTC...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .............................................................................................................................................................................................................TGTAGAATAGAGATGAACAGACCATTCATGTT................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ................................................................................................................................................................................................TGAGAGCTAGGCCTGTAGAATAGAGATGAAC............................................................................................................................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................GTAGAATAGAGATGAACAGACCATTCATG................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ........................................TAATCTATAGTTTTGA...................................................................................................................................................................................................................................................................................................... | 16 | 2 | 0.50 | 0.50 | - | 0.50 | - | - |
| GTTTCTTTTAGTCTTCAGATATTTTGTTCTGGTCGGTCCCTAATCTATAGTTTTGAAACTCGTGCCGCACTTGAAAGGGAAACATCCTACTACCTTTCTTTGCTCTCCATTTCCCACAAACCCTTTCCTTCATCGTCACTCCCTCCATTCTTCTCAGCATATAGATACAAGACCCTTCTTTCTATTATTTGTTGAGAGCTAGGCCTGTAGAATAGAGATGAACAGACCATTCATGTTTCCCTGAAGCTATTTAATGCTGCTCCCAGTCTTCTGTCCATACCATTCCTTCCCTGCCCCTAGGAGTGGGGTTAAGGTCTGTGACATGCAGACTGTAAGCACAGTCAGAATCT .........................................................................................................................................................................................................(((.....((((....(((.(((((........(((((...)))))........((((....))))..))))).)))...)))).......)))....................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553581(SRX182787) source: Testis. (testes) | SRR553607(SRX182813) source: Testis. (testes) | SRR553606(SRX182812) source: Kidney. (Kidney) | SRR553579(SRX182785) source: Heart. (Heart) |
|---|