| (1) BRAIN | (1) HEART | (1) KIDNEY | (2) OTHER |
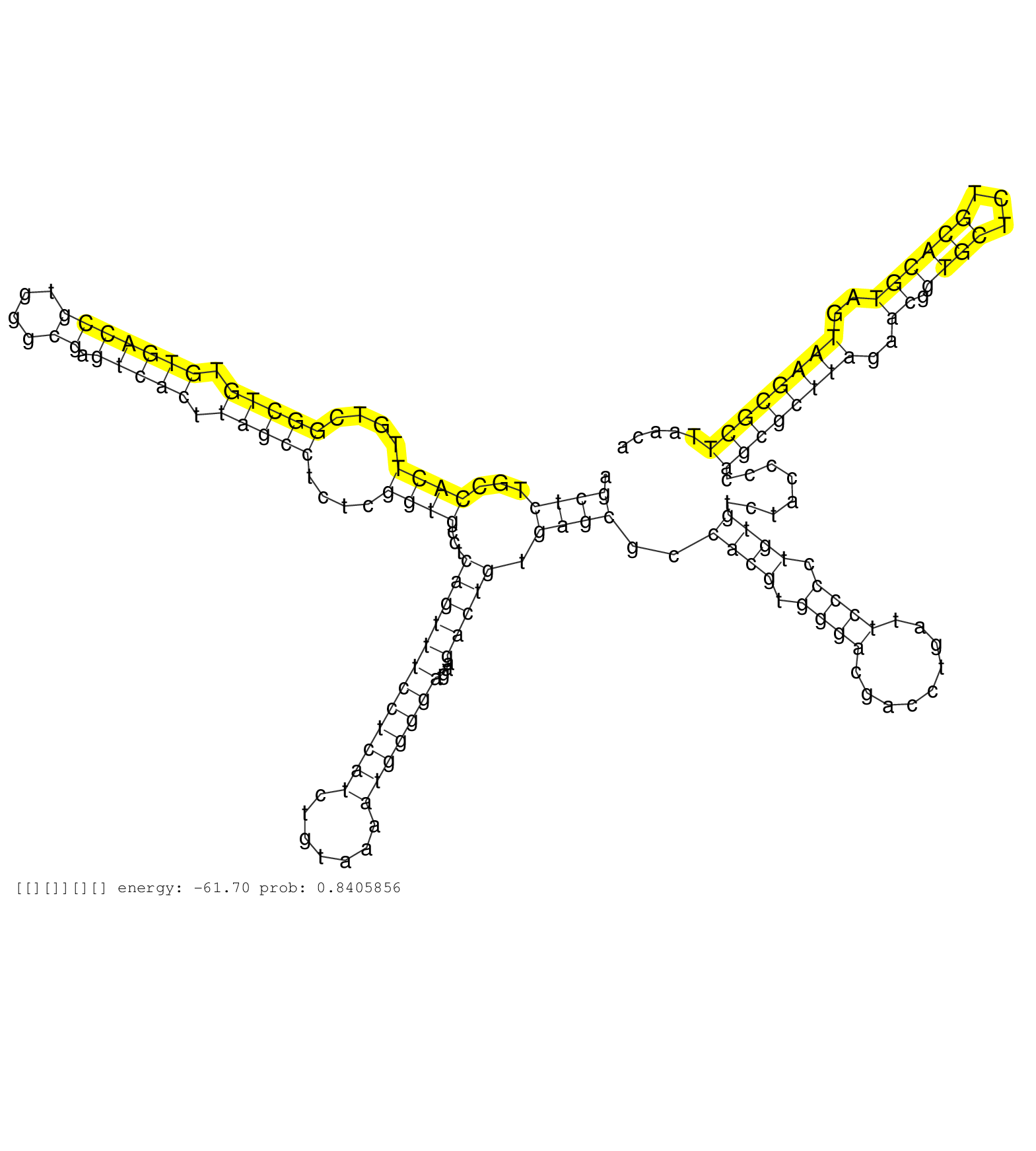
| AAGGAGGTCTGACCCTCTCTTTACATAGAAGCAGCGTGGCTCTAGTGGAAAGAGCCCGGGCTTGGGAGTCAGGGGTCGTGGGTTCAAATCCCAGCTCTGCCACTTGTCGGCTGTGTGACCGTGGGCGAGTCACTTAGCCTCTCGGTGCCTCAGTTTCCTCATCTGTAAAATGGGGATGAAGACTGTGAGCGCCACGTGGGACGACCTGATTCCCCTGTGTCTACCCCAGCGCTTAGAACGGTGCTCTGCACGTAGTAAGCGCTTAACAAATACCGACATTATTATCATTATTGTTATTATTACAGAGGAGAATCCTGCCTAAGCCGACGAGAAAAAGCCGCACCAAGAAC .............................................................................................((((...((((....(((((.(((((((....)).))))).)))))....))))...((((((((((((.......)))))))....))))).))))..((((.((((.........)))).))))........((((((((..((.((((...))))))..))))))))....................................................................................... ............................................................................................93.............................................................................................................................................................................268................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................TGCCACTTGTCGGCTGTGTGACCGAAA.................................................................................................................................................................................................................................. | 27 | 2.00 | 0.00 | 2.00 | - | - | - | - | |
| .................................................................................................................................................................................................................................................TGCTCTGCACGTAGTAAGCGAGA...................................................................................... | 23 | 2.00 | 0.00 | - | 2.00 | - | - | - | |
| .....................................................................................................................................................................................................................................CGCTTAGAACGGTGCTCTGAATA.................................................................................................. | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| ...................................................................................................................................................................................................................................................................................................................GAGAATCCTGCCTAAGCCGACGA.................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................................................................................................................................................GGAGAATCCTGCCTAAGCCGACGAGAAAAAGC............ | 32 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................TAAGCCGACGAGAAAAAGCC........... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 |
| .................................................................................................TGCCACTTGTCGGCTGTGTGAAA...................................................................................................................................................................................................................................... | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| ...............................................................................................................TGTGTGACCGTGGGCGAGTAAA......................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | |
| ..................................................................................................................................................................................................................................................................................................................GGAGAATCCTGCCTAAGCCGACGA.................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - |
| ..............................................................................................................CTGTGTGACCGTGGGCGAGAAC.......................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | 1.00 | - | - | |
| ..................................................................................................................................................................................................................................................................................................................GGAGAATCCTGCCTAAGCCGACGAG................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 |
| ..................................................................................................................................................................................................................................................................................................................GGAGAATCCTGCCTAAGCCGAC...................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - |
| .................................................................................................................................................................................................................................................................................................................................AGCCGACGAGAAAAAGCCGCACCA..... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ....................................................................................................................................................................................................TGGGACGACCTGATTCCCCTGTGTAAA............................................................................................................................... | 27 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| ..............................................................................................................CTGTGTGACCGTGGGCGAGAAGA......................................................................................................................................................................................................................... | 23 | 1.00 | 0.00 | - | - | 1.00 | - | - |
| AAGGAGGTCTGACCCTCTCTTTACATAGAAGCAGCGTGGCTCTAGTGGAAAGAGCCCGGGCTTGGGAGTCAGGGGTCGTGGGTTCAAATCCCAGCTCTGCCACTTGTCGGCTGTGTGACCGTGGGCGAGTCACTTAGCCTCTCGGTGCCTCAGTTTCCTCATCTGTAAAATGGGGATGAAGACTGTGAGCGCCACGTGGGACGACCTGATTCCCCTGTGTCTACCCCAGCGCTTAGAACGGTGCTCTGCACGTAGTAAGCGCTTAACAAATACCGACATTATTATCATTATTGTTATTATTACAGAGGAGAATCCTGCCTAAGCCGACGAGAAAAAGCCGCACCAAGAAC .............................................................................................((((...((((....(((((.(((((((....)).))))).)))))....))))...((((((((((((.......)))))))....))))).))))..((((.((((.........)))).))))........((((((((..((.((((...))))))..))))))))....................................................................................... ............................................................................................93.............................................................................................................................................................................268................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................aagACACAGCCGACAAGTGGCAGGAA....................................................................................................................................................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| ................................................................................................................................................................................................................................................................aagTCGGTATTTGTTAAGCGGAA....................................................................... | 23 | 1.00 | 0.00 | - | - | - | 1.00 | - |