| (1) HEART | (2) OTHER |
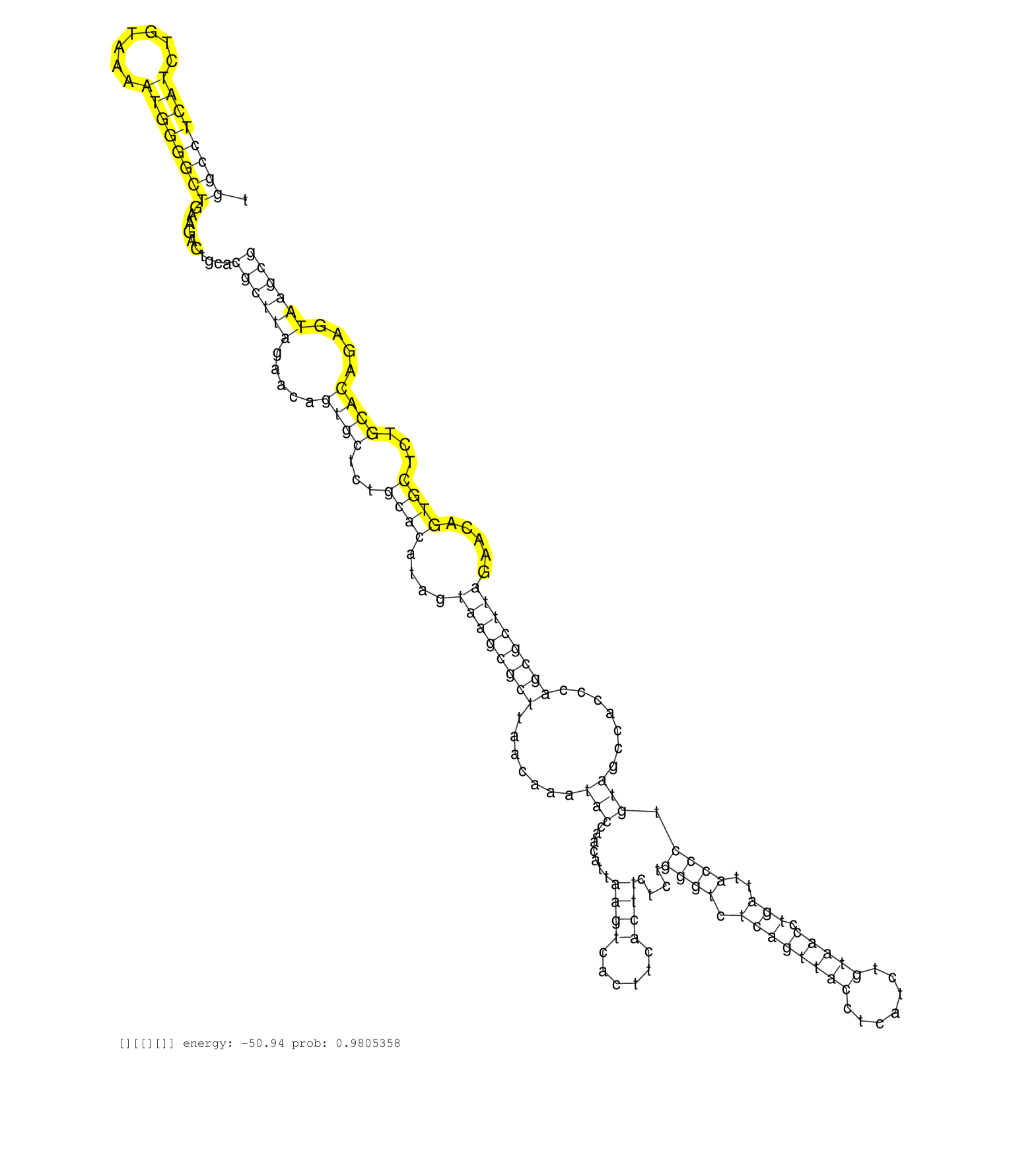
| AGTTCAAATCCCGGCTCTGCCACTTGTCCGCTGTGTGTCTGTGGGCACGTCACTTCACTTCTCTGGGCCTCAGTGGCCTCATCTGTAAAATGGGGCTGAAGACTGCACGCTTAGAACAGTGCTCTGCACATAGTAAGCGCTTAACAAATACCAACATTAAGTCACTTCACTTCTCTGGGTCTCAGTTACCTCATCTGTAACCTGATTACCCTGTAGCCACCCAGCGCTTAGAACAGTGCTCTGCACAGAGTAAGCGCTTAACAGAAGCCAACATCGTGATTATTATTCCCTTCCCACCAGACAAAGTGAATAATGTCACCGTGAAGGTCATCGACCAGACCCGGATGGAT ..........................................................................((((((((.......))))))))..........((((((.....((((...((((....((((((((.......(((.......((((......))))....((((.((((((((.......))))).)))..)))).))).......)))))))).....))))...))))....)))))).............................................................................................. .........................................................................74....................................................................................................................................................................................256............................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................................TTAGAACAGTGCTCTGCACAGAGATT................................................................................................. | 26 | 2.00 | 0.00 | 2.00 | - | - | |
| ................................................................................................................................................................................................................................CGCTTAGAACAGTGCTCTGCACAGATGAC................................................................................................. | 29 | 1.00 | 0.00 | 1.00 | - | - | |
| ..............................................................................TCATCTGTAAAATGGGGCTGAGCTT....................................................................................................................................................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - | |
| ......................................................................................................................................................................................................................................GAACAGTGCTCTGCACAGATGG.................................................................................................. | 22 | 1.00 | 0.00 | - | 1.00 | - | |
| ...................................................................................................................................................................................................................................TTAGAACAGTGCTCTGCACAGATGGC................................................................................................. | 26 | 1.00 | 0.00 | - | 1.00 | - | |
| ....................................................................................................................................................................................................................................TAGAACAGTGCTCTGCACAGAGAAGA................................................................................................ | 26 | 1.00 | 0.00 | 1.00 | - | - | |
| ................................................................................ATCTGTAAAATGGGGCTGACGC........................................................................................................................................................................................................................................................ | 22 | 1.00 | 0.00 | 1.00 | - | - | |
| .....................................................................................................................................................................................................................................AGAACAGTGCTCTGCACAGATGG.................................................................................................. | 23 | 1.00 | 0.00 | - | 1.00 | - | |
| ...................................................................................TGTAAAATGGGGCTGAAGACTGCT................................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| ..................................................................................CTGTAAAATGGGGCTGAAGATCGT.................................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| ......................................................................................................................................................................................................................................GAACAGTGCTCTGCACAGATGCA................................................................................................. | 23 | 1.00 | 0.00 | - | 1.00 | - | |
| ..............................................................................TCATCTGTAAAATGGGGCTGAAGACTGC.................................................................................................................................................................................................................................................... | 28 | 11 | 0.18 | 0.18 | 0.18 | - | - |
| .................................................................................TCTGTAAAATGGGGCTGAAGACTGC.................................................................................................................................................................................................................................................... | 25 | 16 | 0.06 | 0.06 | 0.06 | - | - |
| AGTTCAAATCCCGGCTCTGCCACTTGTCCGCTGTGTGTCTGTGGGCACGTCACTTCACTTCTCTGGGCCTCAGTGGCCTCATCTGTAAAATGGGGCTGAAGACTGCACGCTTAGAACAGTGCTCTGCACATAGTAAGCGCTTAACAAATACCAACATTAAGTCACTTCACTTCTCTGGGTCTCAGTTACCTCATCTGTAACCTGATTACCCTGTAGCCACCCAGCGCTTAGAACAGTGCTCTGCACAGAGTAAGCGCTTAACAGAAGCCAACATCGTGATTATTATTCCCTTCCCACCAGACAAAGTGAATAATGTCACCGTGAAGGTCATCGACCAGACCCGGATGGAT ..........................................................................((((((((.......))))))))..........((((((.....((((...((((....((((((((.......(((.......((((......))))....((((.((((((((.......))))).)))..)))).))).......)))))))).....))))...))))....)))))).............................................................................................. .........................................................................74....................................................................................................................................................................................256............................................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................................taatGTTAAGCGCTTACTCTGTAAT.................................................................................... | 25 | 1.00 | 0.00 | - | - | 1.00 | |
| .............................................................................................................................................................................................................................................attTGTTAAGCGCTTACTCTGTGCAGTTA.................................................................................... | 29 | 1.00 | 0.00 | 1.00 | - | - |