| (1) BRAIN | (1) HEART | (1) KIDNEY | (2) OTHER |
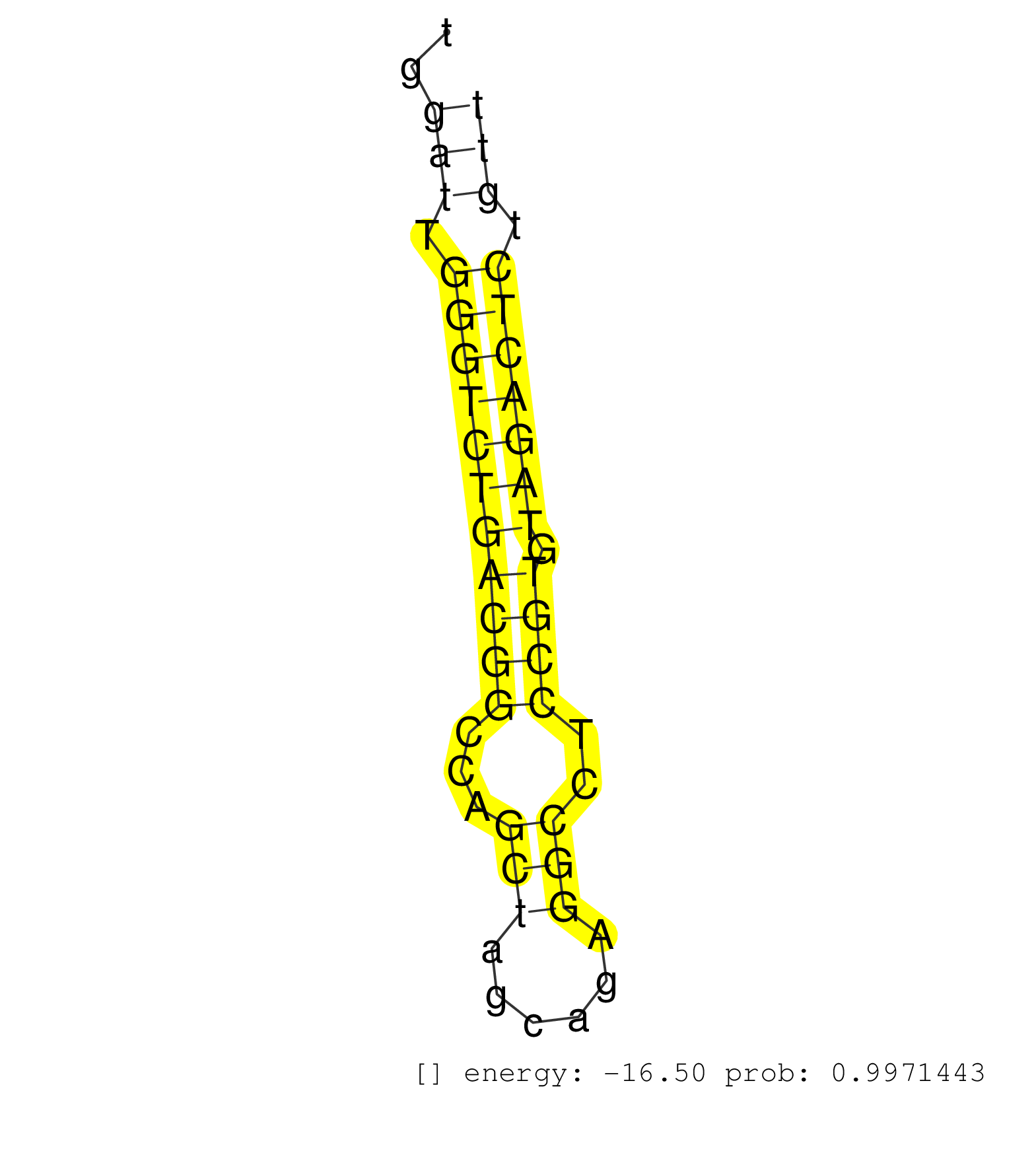
| GAAAACACGGCTTATTAATCAGGTGTTGGAACTTCAGCACACACTTGAAGGTTAGTTTGCTTTTAATAATTGGCCCTACAAAGGGCTAGTTGGACTGGTGACATTTGGGATGCAGAAACAGTATAAACTCCCACTAGAGAAACCGACTCCATTACGGTAACTGTAAACTGGAGCGTCTGCTTGGATTGGGTCTGACGGCCAGCTAGCAGAGGCCTCCGTGTAGACTCTGTTTAATCCAGATAGGAGTCTTGGGTACTCCATTTGGGCCTCTTGGTTCTCTCTGTAGCCTCTCCCCTCTGTTATGAGAACTAGTTCCTTCGAGTCCCCAGATCGTGACCGTTCTACCTTCA .......................................................................................................................................................................................(((.(((((((((((...(((......)))..)))).))))))).)))....................................................................................................................... .....................................................................................................................................................................................182..............................................231..................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................TTGGGTCTGACGGCC...................................................................................................................................................... | 15 | 3 | 3.67 | 3.67 | 0.67 | 0.67 | 1.00 | 0.67 | 0.67 |
| ..........................................................................................................................................................................................TGGGTCTGACGGCCATC................................................................................................................................................... | 17 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| ....................AGGTGTTGGAACTTCAGCA....................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..................TCAGGTGTTGGAACTTC........................................................................................................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - |
| ..........CTTATTAATCAGGTGTTGGAACTTC........................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ........................GTTGGAACTTCAGCACACACTTGAAG............................................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| .................................................................................................................................................................................................................AGGCCTCCGTGTAGACTC........................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................................TGGGTCTGACGGCCAGCT.................................................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - |
| .................ATCAGGTGTTGGAACT............................................................................................................................................................................................................................................................................................................................. | 16 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - |
| .................ATCAGGTGTTGGAAC.............................................................................................................................................................................................................................................................................................................................. | 15 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - |
| GAAAACACGGCTTATTAATCAGGTGTTGGAACTTCAGCACACACTTGAAGGTTAGTTTGCTTTTAATAATTGGCCCTACAAAGGGCTAGTTGGACTGGTGACATTTGGGATGCAGAAACAGTATAAACTCCCACTAGAGAAACCGACTCCATTACGGTAACTGTAAACTGGAGCGTCTGCTTGGATTGGGTCTGACGGCCAGCTAGCAGAGGCCTCCGTGTAGACTCTGTTTAATCCAGATAGGAGTCTTGGGTACTCCATTTGGGCCTCTTGGTTCTCTCTGTAGCCTCTCCCCTCTGTTATGAGAACTAGTTCCTTCGAGTCCCCAGATCGTGACCGTTCTACCTTCA .......................................................................................................................................................................................(((.(((((((((((...(((......)))..)))).))))))).)))....................................................................................................................... .....................................................................................................................................................................................182..............................................231..................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................................CTGGATTAAACAGAG............................................................................................................... | 15 | 7 | 0.14 | 0.14 | - | 0.14 | - | - | - |