| (1) HEART | (1) KIDNEY | (1) OTHER |
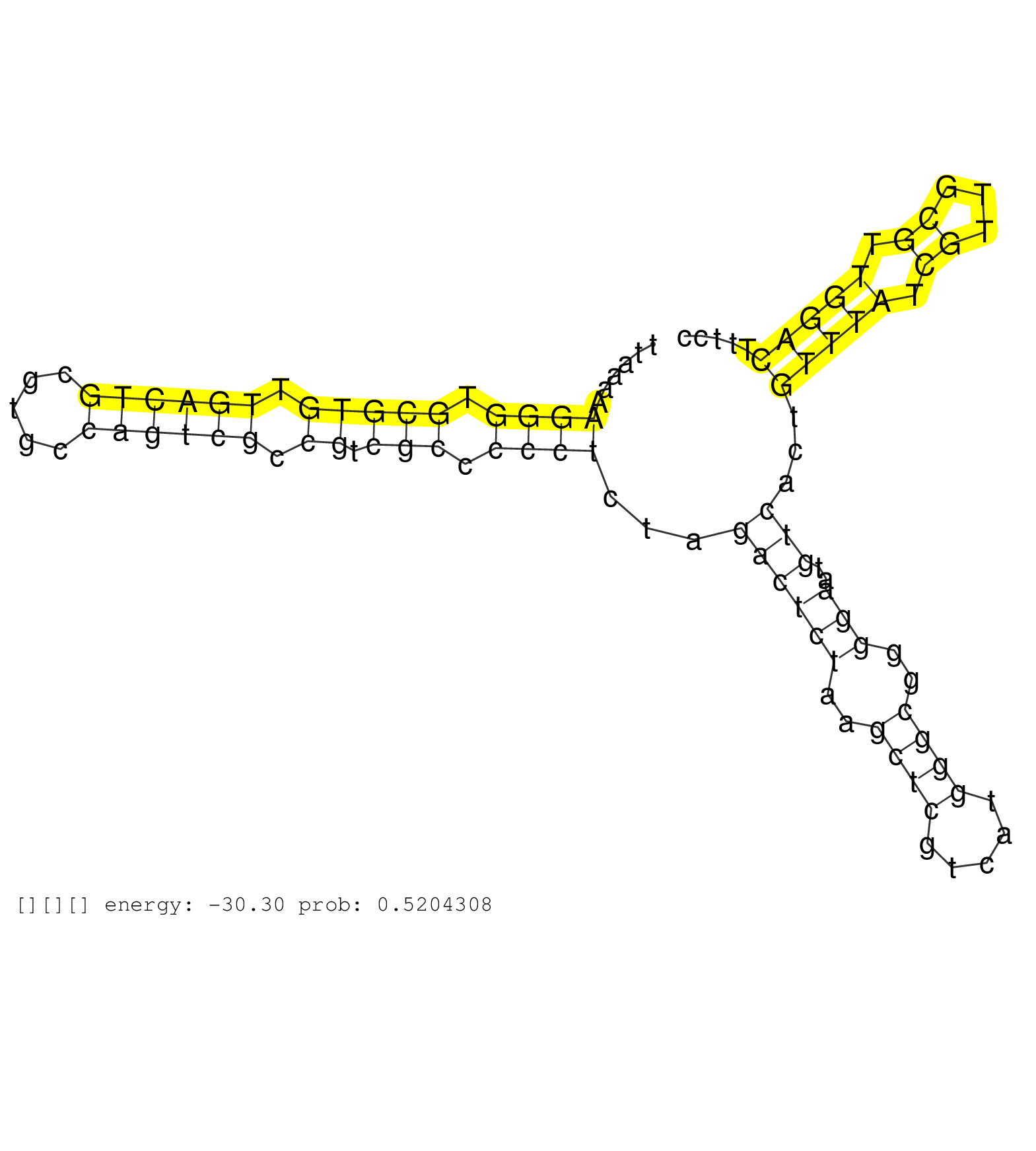
| CCCGCTGGAGACCAGAAACATCGCCTTCTTCTCCACCAACTGCGTCGAAGGTGAGACCCTGCCCGGCGGGGCTGATGATTCCTAATCGTCGTGCTGTTATATTCTTCTTAAAAAGGGTGCGTGTTGACTGCGTGCCAGTCGCCGTCGCCCCCTCTAGACTCTAAGCTCGTCATGGGCGGGGAATGTCACTGTTTATCGTTGCGTTGGACTTTCCCAAGCGCTCAAAAGAATGCTGTGCACACAGTAAGCGGTCAAGAGAAACGACCGAATGAAGGAATTTCTGAAGGTCCTCCCCTCGCCAGCCTGCGGTCTTTGCTCTAGGCCGTGTTGCTTCTCAGAGGGGTGAGGGA .................................................................................................................((((.(((((.((((((.....)))))).)).))).))))...((((((..((((.....))))..)))..)))...(((((.((...)).)))))............................................................................................................................................. ...........................................................................................................108.......................................................................................................214...................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|
| ..................................ACCAACTGCGTCGAAGGAAC........................................................................................................................................................................................................................................................................................................ | 20 | 1.00 | 0.00 | - | 1.00 | - | |
| .................................CACCAACTGCGTCGAAGGAAC........................................................................................................................................................................................................................................................................................................ | 21 | 1.00 | 0.00 | - | 1.00 | - | |
| ...............AAACATCGCCTTCTTCTCCACCAAC...................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...........................CTTCTCCACCAACTGCGTCG............................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ...................................................................................................................................................................................................................................................................AACGACCGAATGAAGGAATTTCTGAAG................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ....................................................................................................................................................................................................................................AATGCTGTGCACACAGTAAGCGGTCAAG.............................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..............................................................................................................................................................................................................................................................................GAAGGAATTTCTGAAGGTCCTCCCCTCGCC.................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ................................................................................................................AAGGGTGCGTGTTGACTG............................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .................................CACCAACTGCGTCGAAGGAACG....................................................................................................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | |
| ......GGAGACCAGAAACATCGCCTTCTTC............................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..............................................................................................................................................................................................GTTTATCGTTGCGTTGGACT............................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ........................................................................................................................................................TCTAGACTCTAAGCTCGTCATGGGC............................................................................................................................................................................. | 25 | 8 | 0.25 | 0.25 | - | - | 0.25 |
| ..........................................................................................................................................................TAGACTCTAAGCTCGTCATGGGC............................................................................................................................................................................. | 23 | 8 | 0.25 | 0.25 | - | - | 0.25 |
| ..........................................................................................................................................................TAGACTCTAAGCTCGTCATGGG.............................................................................................................................................................................. | 22 | 8 | 0.25 | 0.25 | - | - | 0.25 |
| ...........................................................................................................................................................AGACTCTAAGCTCGTCATGGG.............................................................................................................................................................................. | 21 | 9 | 0.11 | 0.11 | 0.11 | - | - |
| CCCGCTGGAGACCAGAAACATCGCCTTCTTCTCCACCAACTGCGTCGAAGGTGAGACCCTGCCCGGCGGGGCTGATGATTCCTAATCGTCGTGCTGTTATATTCTTCTTAAAAAGGGTGCGTGTTGACTGCGTGCCAGTCGCCGTCGCCCCCTCTAGACTCTAAGCTCGTCATGGGCGGGGAATGTCACTGTTTATCGTTGCGTTGGACTTTCCCAAGCGCTCAAAAGAATGCTGTGCACACAGTAAGCGGTCAAGAGAAACGACCGAATGAAGGAATTTCTGAAGGTCCTCCCCTCGCCAGCCTGCGGTCTTTGCTCTAGGCCGTGTTGCTTCTCAGAGGGGTGAGGGA .................................................................................................................((((.(((((.((((((.....)))))).)).))).))))...((((((..((((.....))))..)))..)))...(((((.((...)).)))))............................................................................................................................................. ...........................................................................................................108.......................................................................................................214...................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|