| (1) HEART | (1) OTHER |
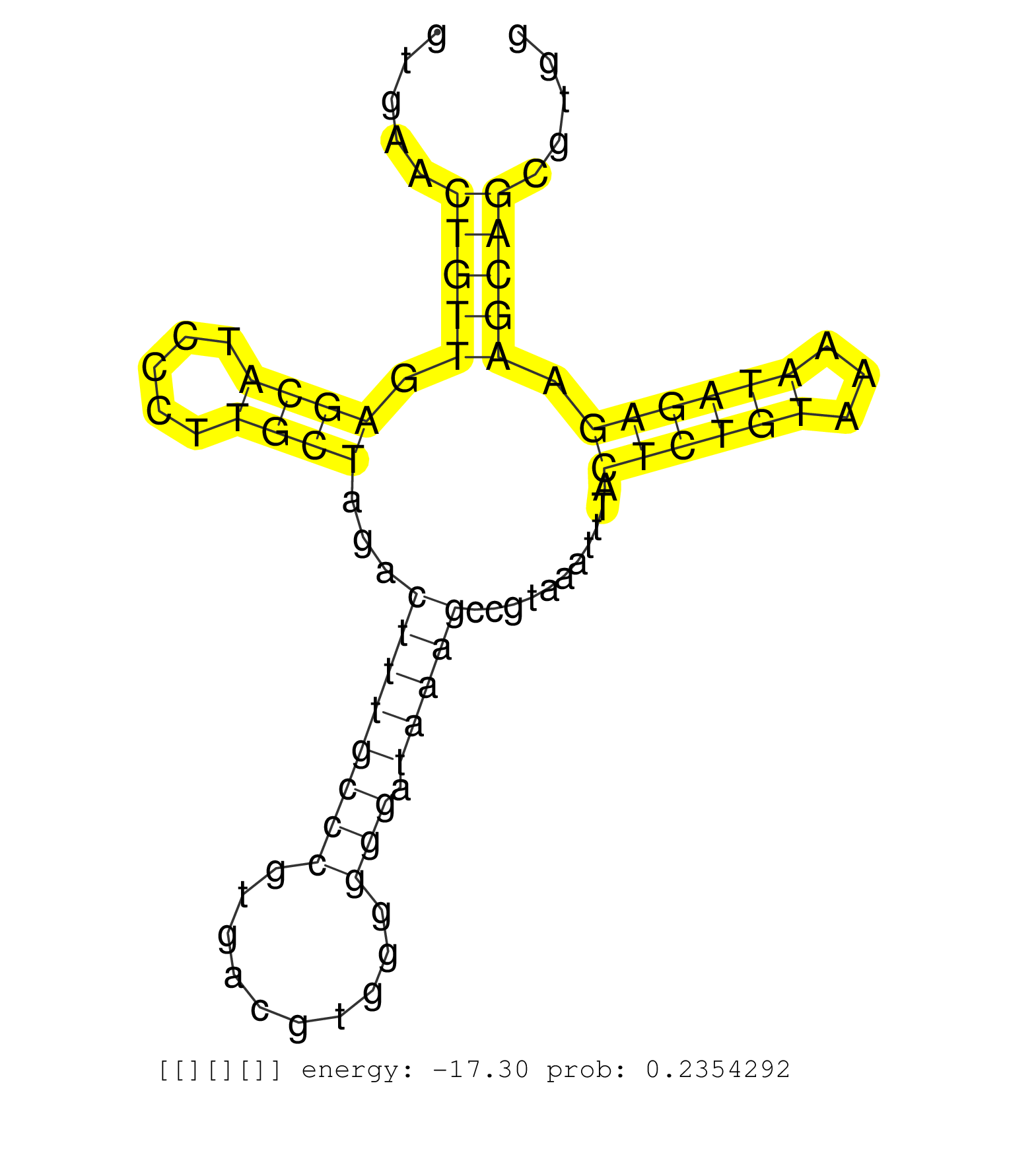
| GACAAAACCAGGATTTACAGAGCCAGGGCAGCTGTTCCCCAGAAACGCAAGTGAACTGTTGAGCATCCCTTGCTAGACTTTGCCCGTGACGTGGGGGGATAAAGCCGTAAATTTACTCTGTAAAATAGAGAAGCAGCGTGGCTCGGTGGAAAGAGCCCGGGCTTTGGAGTCAGAGGTCATGGGTTCGAATCCCAGCTCCGCCACTTGTCAGCTGTGTGACTGTGGGCAAGTCTCTTAACTTCTCTGTGCCTCAGTTCTCATCTGTAAAATGGGGATTAAGACTGTGAACCTCACGTGGGACAACCTGATTACTCTGTGATCTACCCCAGCGCTTAGAACAGTGCTCTGCA .......................................................(((((.((((.....))))...((((((((..........))).)))))...........((((((...)))))).)))))...................................................................................................................................................................................................................... ..................................................51........................................................................................141............................................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|
| .................................................................................................................TACTCTGTAAAATAGAGAAGCAGC..................................................................................................................................................................................................................... | 24 | 1 | 2.00 | 2.00 | 2.00 | - |
| .............................................................................................................................................................................................................................................................................TGGGGATTAAGACTGTGAACCTCCACA...................................................... | 27 | 1.00 | 0.00 | 1.00 | - | |
| ..................................................................................................................................................................TTTGGAGTCAGAGGTCATGGGTTATT.................................................................................................................................................................. | 26 | 1.00 | 0.00 | 1.00 | - | |
| .......................................................................................................................................................................................................................................................................................................TGGGACAACCTGATTACTCTGTGATA............................. | 26 | 1.00 | 0.00 | 1.00 | - | |
| ...............................................................................................................TTTACTCTGTAAAATAGAGAAGCAGCGTG.................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - |
| .....................................................AACTGTTGAGCATCCCTTGCT.................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - |
| ...............................................................................................................................................................................................................................................................................GGGATTAAGACTGTGAACCTCAGTG...................................................... | 25 | 1.00 | 0.00 | - | 1.00 | |
| ...............................................................................................................TTTACTCTGTAAAATAGAGAAGCAGC..................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - |
| ..................................................................................................................................................................TTTGGAGTCAGAGGTCATGGGTTAAAT................................................................................................................................................................. | 27 | 1.00 | 0.00 | 1.00 | - | |
| .........................................................................................................................................................................................................................................................................................................GGACAACCTGATTACTCTGTGATA............................. | 24 | 1.00 | 0.00 | 1.00 | - | |
| ..............................................................................................................................................TCGGTGGAAAGAGCCCGGGCTTTTTT...................................................................................................................................................................................... | 26 | 1.00 | 0.00 | 1.00 | - | |
| ..........................................................................................................................................................................................................................................TTAACTTCTCTGTGCCTCAGTTCTCATC........................................................................................ | 28 | 15 | 0.07 | 0.07 | 0.07 | - |
| GACAAAACCAGGATTTACAGAGCCAGGGCAGCTGTTCCCCAGAAACGCAAGTGAACTGTTGAGCATCCCTTGCTAGACTTTGCCCGTGACGTGGGGGGATAAAGCCGTAAATTTACTCTGTAAAATAGAGAAGCAGCGTGGCTCGGTGGAAAGAGCCCGGGCTTTGGAGTCAGAGGTCATGGGTTCGAATCCCAGCTCCGCCACTTGTCAGCTGTGTGACTGTGGGCAAGTCTCTTAACTTCTCTGTGCCTCAGTTCTCATCTGTAAAATGGGGATTAAGACTGTGAACCTCACGTGGGACAACCTGATTACTCTGTGATCTACCCCAGCGCTTAGAACAGTGCTCTGCA .......................................................(((((.((((.....))))...((((((((..........))).)))))...........((((((...)))))).)))))...................................................................................................................................................................................................................... ..................................................51........................................................................................141............................................................................................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................................................................cggcTACAGATGAGACGGC................................................................................ | 19 | 1.00 | 0.00 | 1.00 | - | |
| .................................................................................................................................................................................................................................................gacGATGAGAACTGAGGCACACAG..................................................................................... | 24 | 1.00 | 0.00 | - | 1.00 | |
| ....................................................................................................................................................................................................................................................tTTACAGATGAGAACTGAGGCACT.................................................................................. | 24 | 1.00 | 0.00 | 1.00 | - |