| (1) HEART | (1) KIDNEY | (1) OTHER |
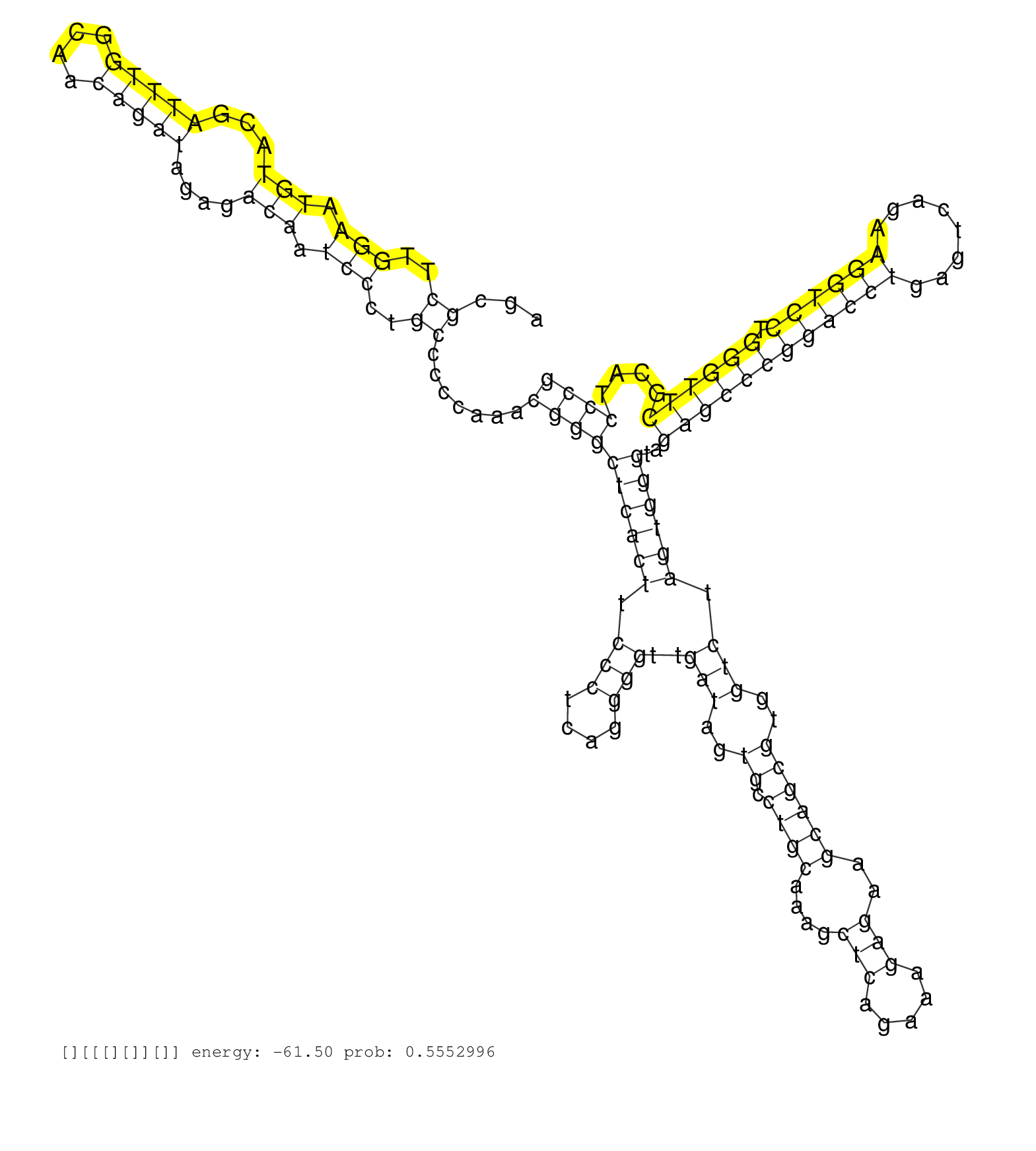
| ACGTGTGGAGTACCATTTTAAGCCCTGAGGTAGATTCATTCGTTCCTTCGGTCGTATTTATTGAGTGCTTACTATGTACGGAACACTGTACTAAGCGCTTGGAATGTACGATTTGGCAACAGATAGAGACAATCCCTGCCCCCAAACGGGCTCACTTCCCTCAGGGGTTGATAGTGCCTGCAAAGCTCAGAAAGAGAAGCAGCGTGGTCTAGTGGGTAGAGCCCGGACCTGAGTCAGAAGGTCCTGGGTTCGCATCCCGGCTCCGCCGCTTGTCTGCTGCGTGACCTTGGGCAAGTCCCTTCACTTCTCTGGGCCTCAGTTGCCTCAGCCGTAAAAGGGAGATCGAGACT ................................................................................................((..(((.(((...(((((....)))))....))).)))..)).......((((((((((.(((....)))..(((..((.((((....(((.....)))..))))))..))).))))))..((((((((((((........)))))).))))))....))))........................................................................................... .............................................................................................94...................................................................................................................................................................259......................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................................AAGGTCCTGGGTTCGTGC............................................................................................... | 18 | 10.00 | 0.00 | 10.00 | - | - | |
| ............................................................................................................................................................................................................................................GAAGGTCCTGGGTTCGTGC............................................................................................... | 19 | 7.00 | 0.00 | 7.00 | - | - | |
| ............................................................................................................................................................................................................................................GAAGGTCCTGGGTTCGTGCC.............................................................................................. | 20 | 6.00 | 0.00 | 5.00 | 1.00 | - | |
| .............................................................................................................................................................................................................................................AAGGTCCTGGGTTCGTGCC.............................................................................................. | 19 | 6.00 | 0.00 | 5.00 | 1.00 | - | |
| ............................................................................................................................................................................................................................................GAAGGTCCTGGGTTCGCGC............................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | |
| ..................................................................................................TTGGAATGTACGATTTGTTA........................................................................................................................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | 1.00 | |
| ............................................................................................................................................................................................................................................GAAGGTCCTGGGTTCGCGCC.............................................................................................. | 20 | 1.00 | 0.00 | 1.00 | - | - | |
| ...................................................................................................................................................................................................................................................CTGGGTTCGCATCCCAGCG........................................................................................ | 19 | 1.00 | 0.00 | 1.00 | - | - | |
| ...........................................................................................................................................................................................................................................AGAAGGTCCTGGGTTCGCGC............................................................................................... | 20 | 1.00 | 0.00 | - | 1.00 | - | |
| ......................................................................................................................................................................................................................GGTAGAGCCCGGACCGGGC..................................................................................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | |
| ..................................................................................................TTGGAATGTACGATTTGGAAAA...................................................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | 1.00 | |
| ...................................................................................................TGGAATGTACGATTTGGCAACATACA................................................................................................................................................................................................................................. | 26 | 1.00 | 0.00 | - | - | 1.00 | |
| .................................................................................................CTTGGAATGTACGATTTGGCAACCT.................................................................................................................................................................................................................................... | 25 | 1.00 | 0.00 | - | 1.00 | - | |
| ..............................................................................................................................................................................................................................................AGGTCCTGGGTTCGCGCC.............................................................................................. | 18 | 1.00 | 0.00 | - | 1.00 | - | |
| .............................................................................................................................................................................................................................................AAGGTCCTGGGTTCGC................................................................................................. | 16 | 2 | 0.50 | 0.50 | 0.50 | - | - |
| .....................................................................................................................AACAGATAGAGACAATCCCTGCCCCCAAAC........................................................................................................................................................................................................... | 30 | 11 | 0.09 | 0.09 | - | 0.09 | - |
| ACGTGTGGAGTACCATTTTAAGCCCTGAGGTAGATTCATTCGTTCCTTCGGTCGTATTTATTGAGTGCTTACTATGTACGGAACACTGTACTAAGCGCTTGGAATGTACGATTTGGCAACAGATAGAGACAATCCCTGCCCCCAAACGGGCTCACTTCCCTCAGGGGTTGATAGTGCCTGCAAAGCTCAGAAAGAGAAGCAGCGTGGTCTAGTGGGTAGAGCCCGGACCTGAGTCAGAAGGTCCTGGGTTCGCATCCCGGCTCCGCCGCTTGTCTGCTGCGTGACCTTGGGCAAGTCCCTTCACTTCTCTGGGCCTCAGTTGCCTCAGCCGTAAAAGGGAGATCGAGACT ................................................................................................((..(((.(((...(((((....)))))....))).)))..)).......((((((((((.(((....)))..(((..((.((((....(((.....)))..))))))..))).))))))..((((((((((((........)))))).))))))....))))........................................................................................... .............................................................................................94...................................................................................................................................................................259......................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|
| ............................................gtagCAATAAATACGACCGGATG........................................................................................................................................................................................................................................................................................... | 23 | 1.00 | 0.00 | - | - | 1.00 | |
| .....................................ACCGAAGGAACGAAT.......................................................................................................................................................................................................................................................................................................... | 15 | 19 | 0.05 | 0.05 | - | 0.05 | - |