| (1) HEART | (1) KIDNEY | (2) OTHER |
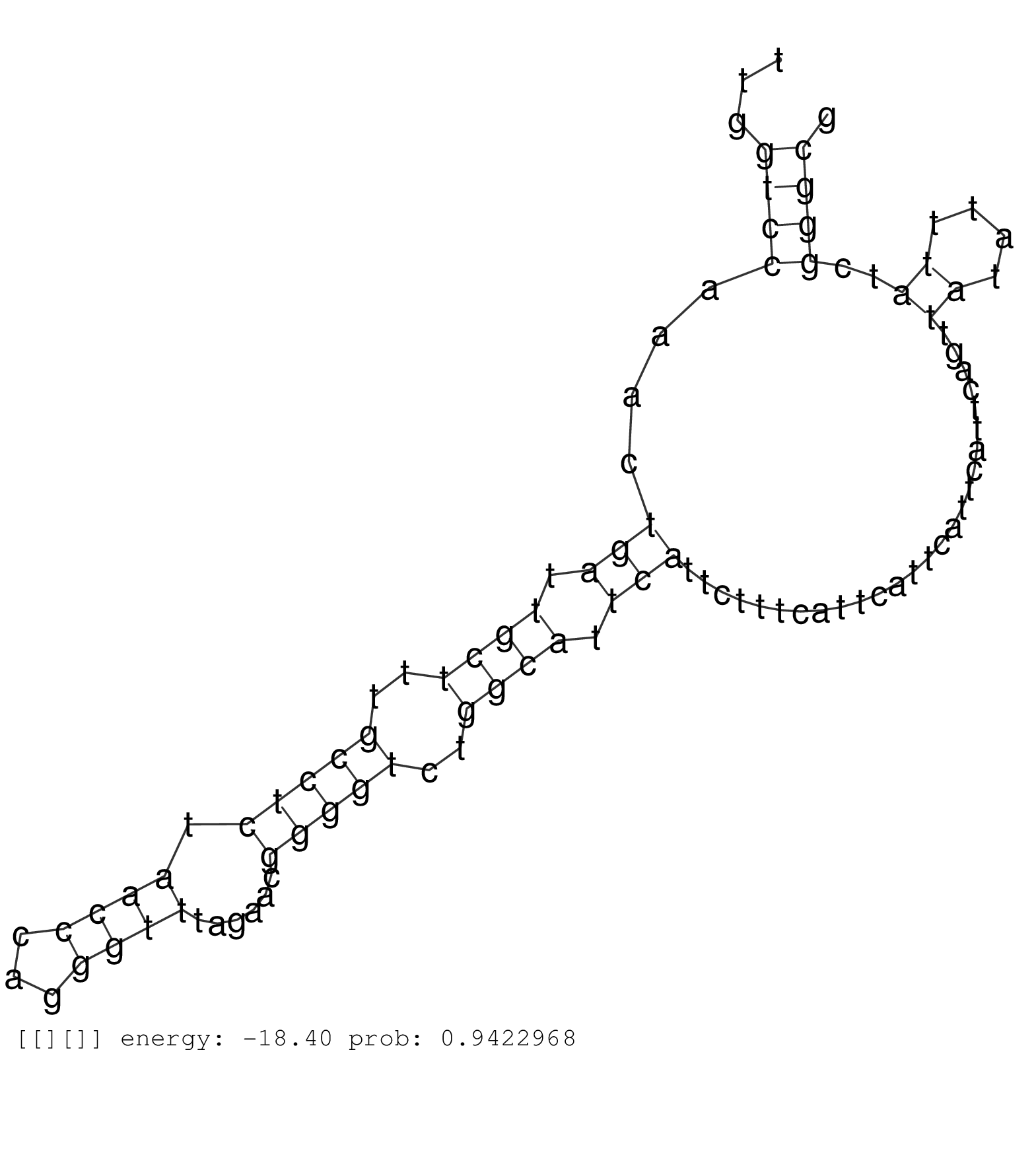
| GCGGACATGTTCGCGAAGGATAGAGACACGCTGGACTTCATCAGGAGACTGTAGGTGGCCCCTGGGACTCCAAAGCGGGTGATGGGTGGCGTCGGGACCTAGCTGTGGGACCTGGGGCGAGTCACTTAACTTCTCTGTGCCTTGATTATCTCATCTGTAAAAGGGGAATTAAGACTGTGAACCGTGCGTGGGACCCGGACTTGGTCCAAACTGATTGCTTTGCCTCTAACCCAGGGTTTAGAACGGGGTCTGGCATTCATTCTTTCATTCATTCATTCATTCAGTTATATTTATCGGGCGCTTACTGGGTGGAGCGGGTGGGCAGACACCCCCTGCGCCTCTCCTGCCTC ...........................................................................................................................................................................................................((((....(((.((((..(((((.((((...))))......)))))..)))).)))..........................((....))..))))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................TCTGTAAAAGGGGAATTAAGACTGTTAGC........................................................................................................................................................................ | 29 | 4.00 | 0.00 | 4.00 | - | - | - | |
| .............................................................................................................................................................TAAAAGGGGAATTAAGACTGTAGCC........................................................................................................................................................................ | 25 | 2.00 | 0.00 | 2.00 | - | - | - | |
| ........................................................................................................................................................ATCTGTAAAAGGGGAATTAAGACTGTCAGC........................................................................................................................................................................ | 30 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ..................GATAGAGACACGCTGGACTTCATCAGGA................................................................................................................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..................GATAGAGACACGCTGGACTTCATCAGGAT............................................................................................................................................................................................................................................................................................................... | 29 | 1.00 | 1.00 | - | 1.00 | - | - | |
| ...............AAGGATAGAGACACGCT.............................................................................................................................................................................................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ..................GATAGAGACACGCTGGACTTCAT..................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ......................GAGACACGCTGGACTTCATCAGGAGACTGTGCC....................................................................................................................................................................................................................................................................................................... | 33 | 1.00 | 0.00 | - | - | - | 1.00 | |
| ....ACATGTTCGCGAAGGATAGAGACACGCT.............................................................................................................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ...........................ACGCTGGACTTCATCAGGAGACT............................................................................................................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ......................................................................................................................................................TCATCTGTAAAAGGGGAATTAAGACTGTGTT......................................................................................................................................................................... | 31 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .........TTCGCGAAGGATAGAGACACGCTGGACT......................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ......................................................................................................................................................TCATCTGTAAAAGGGGAATTAAGACTGTTT.......................................................................................................................................................................... | 30 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .........................................................................................................................................................TCTGTAAAAGGGGAATTAAGACTGTCAGC........................................................................................................................................................................ | 29 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .......................AGACACGCTGGACTTCATCAGGAGACT............................................................................................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ....................TAGAGACACGCTGGACTTCATCAGG................................................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..........................CACGCTGGACTTCAT..................................................................................................................................................................................................................................................................................................................... | 15 | 3 | 0.33 | 0.33 | - | 0.33 | - | - |
| ....................................................................................................................................................TCTCATCTGTAAAAGGGGAATTAAGACTGT............................................................................................................................................................................ | 30 | 10 | 0.20 | 0.20 | 0.20 | - | - | - |
| GCGGACATGTTCGCGAAGGATAGAGACACGCTGGACTTCATCAGGAGACTGTAGGTGGCCCCTGGGACTCCAAAGCGGGTGATGGGTGGCGTCGGGACCTAGCTGTGGGACCTGGGGCGAGTCACTTAACTTCTCTGTGCCTTGATTATCTCATCTGTAAAAGGGGAATTAAGACTGTGAACCGTGCGTGGGACCCGGACTTGGTCCAAACTGATTGCTTTGCCTCTAACCCAGGGTTTAGAACGGGGTCTGGCATTCATTCTTTCATTCATTCATTCATTCAGTTATATTTATCGGGCGCTTACTGGGTGGAGCGGGTGGGCAGACACCCCCTGCGCCTCTCCTGCCTC ...........................................................................................................................................................................................................((((....(((.((((..(((((.((((...))))......)))))..)))).)))..........................((....))..))))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................CAGTTTGGACCAAGT......................................................................................................................................... | 15 | 12 | 0.08 | 0.08 | - | - | 0.08 | - |