| (1) HEART | (1) KIDNEY | (2) OTHER |
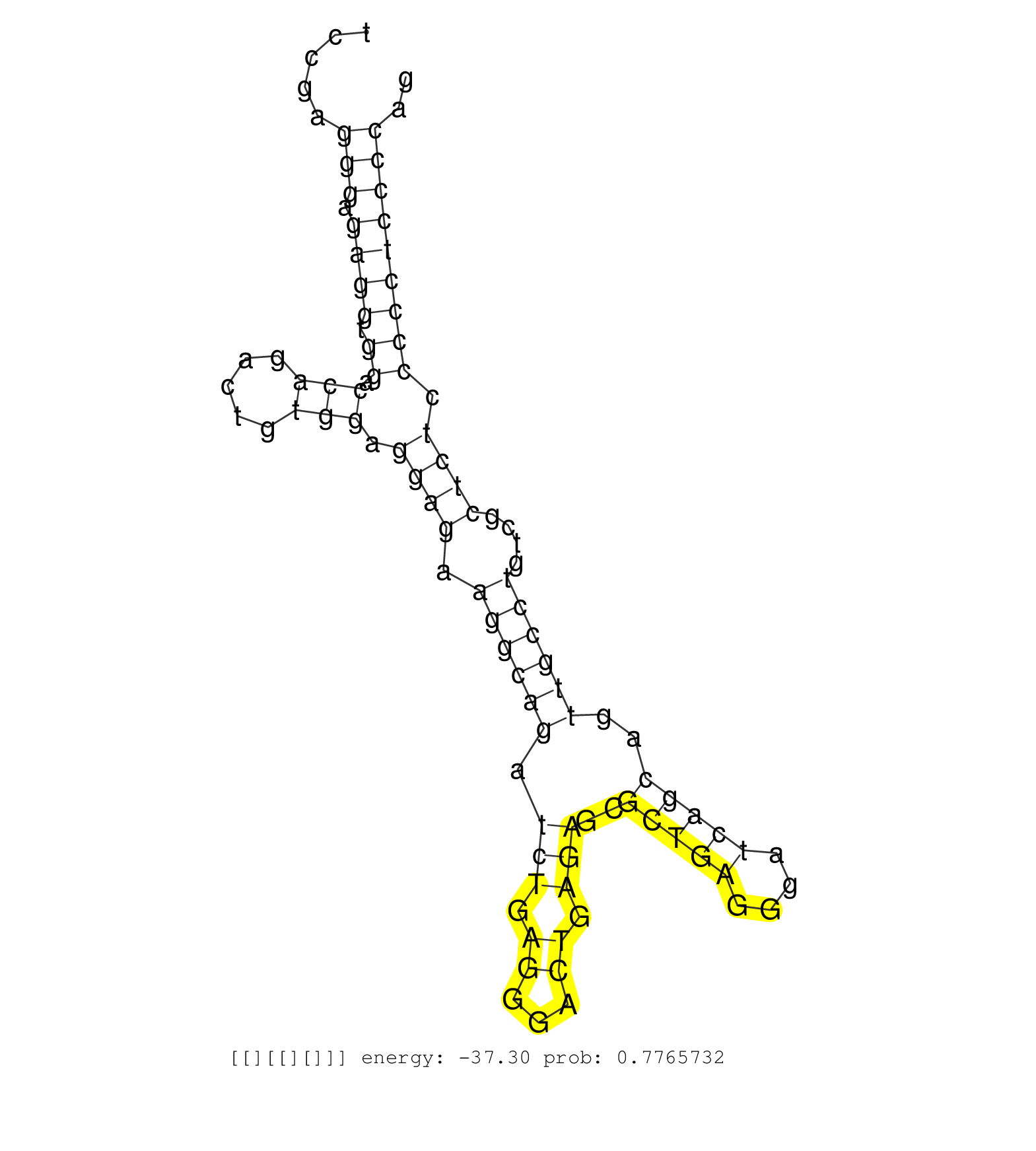
| GCATTTATGTACATATCCATAATTTGTTTTAATGTCTGTCTCCCCCTCTAGACTCTAAGCTCCTGGTGGGCTGGGAATGTGTCTAGCAACTCTGTATCGTACTCTCCCTAGCGTTTAGTACAGTGCTCTGCTGACGGTAACCACTCATTCCACTGATTCGTTGATGGGCAGGGAGGTCAGGGAGGAGACTACTGCAATCCTCCGAGGGAGAGGTGGACCAGACTGTGGAGGAGAAGGCAGATCTGAGGGACTGAGAGCGCTGAGGGATCAGCAGTTGCCTGTCGCTCTCCCCCTCCCCAGATCGTGGGCACCAAGCTGCGCATCCCGCACGTGACCCCGGCCGACTCGGG .............................................................................................................................................................................................................(((.((((.((.(((.....))).((((.((((((.(((.((...)).)))..(((((....)))))..))))))....)))).))))))))).................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................................................TGAGGGACTGAGAGCGCTGAGT..................................................................................... | 22 | 2.00 | 0.00 | - | 1.00 | 1.00 | - | |
| .....................................................................GCTGGGAATGTGTCTAGCAACTCTGTATCGT.......................................................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| .................................................................................................................................................................................................................................................TCTGAGGGACTGAGAGCGCTGA....................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ..................................................................................................................................................................................................................................................CTGAGGGACTGAGAGCGCTGATA..................................................................................... | 23 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ...................................................................................................................................................................................................................................................TGAGGGACTGAGAGCGCTGA....................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ........................................................................................................................................................................................................................................................................................................CCAGATCGTGGGCACCAAGCTGCGC............................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ...................................................................................................................................................................................................................................................TGAGGGACTGAGAGCGCTGTAA..................................................................................... | 22 | 1.00 | 0.00 | - | 1.00 | - | - | |
| .................................................................................................................................................................................................................................................TCTGAGGGACTGAGAGCGCTGAGG..................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ...................................................................................................................................................................................................................................................TGAGGGACTGAGAGCGCTGAGTT.................................................................................... | 23 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ...................................................................................................................................................................................................................................................TGAGGGACTGAGAGCGCTGAGG..................................................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| GCATTTATGTACATATCCATAATTTGTTTTAATGTCTGTCTCCCCCTCTAGACTCTAAGCTCCTGGTGGGCTGGGAATGTGTCTAGCAACTCTGTATCGTACTCTCCCTAGCGTTTAGTACAGTGCTCTGCTGACGGTAACCACTCATTCCACTGATTCGTTGATGGGCAGGGAGGTCAGGGAGGAGACTACTGCAATCCTCCGAGGGAGAGGTGGACCAGACTGTGGAGGAGAAGGCAGATCTGAGGGACTGAGAGCGCTGAGGGATCAGCAGTTGCCTGTCGCTCTCCCCCTCCCCAGATCGTGGGCACCAAGCTGCGCATCCCGCACGTGACCCCGGCCGACTCGGG .............................................................................................................................................................................................................(((.((((.((.(((.....))).((((.((((((.(((.((...)).)))..(((((....)))))..))))))....)))).))))))))).................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|
| .tcggTGGATATGTACATAGGCT....................................................................................................................................................................................................................................................................................................................................... | 22 | 69.00 | 0.00 | 69.00 | - | - | - | |
| .cggTGGATATGTACATAAGGC........................................................................................................................................................................................................................................................................................................................................ | 21 | 27.00 | 0.00 | 27.00 | - | - | - | |
| .ggTGGATATGTACATAAAGG......................................................................................................................................................................................................................................................................................................................................... | 20 | 5.00 | 0.00 | 5.00 | - | - | - | |
| .tcgtTGGATATGTACATATGCT....................................................................................................................................................................................................................................................................................................................................... | 22 | 2.00 | 0.00 | 2.00 | - | - | - | |
| .tctgTGGATATGTACATAGTCT....................................................................................................................................................................................................................................................................................................................................... | 22 | 2.00 | 0.00 | 2.00 | - | - | - | |
| .aaagTGGATATGTACATAGAAA....................................................................................................................................................................................................................................................................................................................................... | 22 | 2.00 | 0.00 | 2.00 | - | - | - | |
| ....gcTATGGATATGTACATCG....................................................................................................................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | 1.00 | |
| ................................................................................................................gtccAGCAGAGCACTGTACTCCTG...................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .aggTGGATATGTACATAAGGA........................................................................................................................................................................................................................................................................................................................................ | 21 | 1.00 | 0.00 | 1.00 | - | - | - |