| (1) HEART | (2) OTHER |
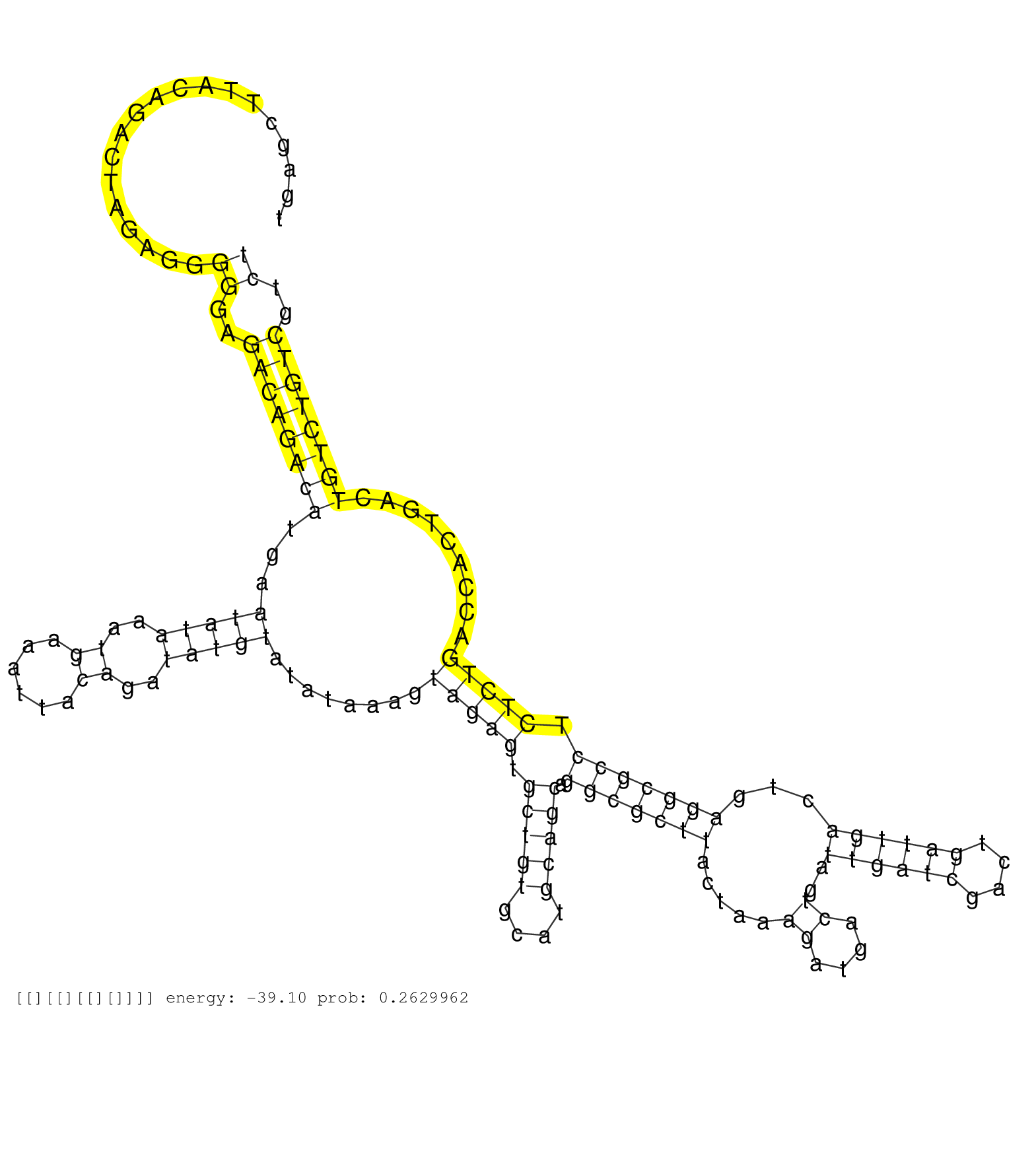
| TCAGCACTCAGTAAAGTGCTCAGCAAGTACACAATAAGCACTTACTAAATAGTCTTACTCAAGTTAAGTCAGTCAGTCGTATTTATCGAGCAAATACAAAGATATAACAGACACCTTCCCTGCCCACAATGAGCTTACAGACTAGAGGGGGAGACAGACATGAATATAAATGAAATTACAGATATGTATATAAAGTAGAGTGCTGTGCATGCAGCAGGCGCTTACTAAAGATGACTGATTGATCGACTGATTGACTGAGGCGCCTCTCTGACCACTGACTGTCTGTCGTCTTGCTCCTAGAATGGCACGTCGGCCCTGCACGCTGCAGTTCTTAGTGGCAACATCCGAGC ....................................................................................................................................................((..((((((((...(((((..((......))..)))))........(((((.(((((....))))).(((((((.....((....))..((((((....))))))...))))))).))))).........))))))))..))........................................................... .................................................................................................................................130..............................................................................................................................................................291......................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| ......................................................................................................................................TTACAGACTAGAGGGGGAGACCGT................................................................................................................................................................................................ | 24 | 14.00 | 0.00 | 14.00 | - | - | |
| ......................................................................................................................................TTACAGACTAGAGGGGGAGACCCAT............................................................................................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - | |
| ........................................................................................................................................................................................................................................................................TCTCTGACCACTGACTGTCTGTC............................................................... | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ......................................................................................................................................TTACAGACTAGAGGGGGAGACCGC................................................................................................................................................................................................ | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| .......................................................................................................................................TACAGACTAGAGGGGGAGACCGT................................................................................................................................................................................................ | 23 | 1.00 | 0.00 | 1.00 | - | - | |
| ......................................................................................................................................TTACAGACTAGAGGGGGAGACCGTT............................................................................................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - | |
| ......................................................................................................................................TTACAGACTAGAGGGGGAGACCGTA............................................................................................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - | |
| ......................................................................................................................................TTACAGACTAGAGGGGGAGACCGAA............................................................................................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - | |
| ......................................................................................................................................TTACAGACTAGAGGGGGAGAACGA................................................................................................................................................................................................ | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| .................................................................................................................................................................................................................................................ATCGACTGATTGACTGAGGCGCC...................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| .......................................................................................................................................................................................................................................................................................................................................GTTCTTAGTGGCAAC........ | 15 | 3 | 0.33 | 0.33 | - | 0.33 | - |
| TCAGCACTCAGTAAAGTGCTCAGCAAGTACACAATAAGCACTTACTAAATAGTCTTACTCAAGTTAAGTCAGTCAGTCGTATTTATCGAGCAAATACAAAGATATAACAGACACCTTCCCTGCCCACAATGAGCTTACAGACTAGAGGGGGAGACAGACATGAATATAAATGAAATTACAGATATGTATATAAAGTAGAGTGCTGTGCATGCAGCAGGCGCTTACTAAAGATGACTGATTGATCGACTGATTGACTGAGGCGCCTCTCTGACCACTGACTGTCTGTCGTCTTGCTCCTAGAATGGCACGTCGGCCCTGCACGCTGCAGTTCTTAGTGGCAACATCCGAGC ....................................................................................................................................................((..((((((((...(((((..((......))..)))))........(((((.(((((....))))).(((((((.....((....))..((((((....))))))...))))))).))))).........))))))))..))........................................................... .................................................................................................................................130..............................................................................................................................................................291......................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................................cctgAGTCAATCAGTCGGTCC.......................................................................................... | 21 | 1.00 | 0.00 | - | 1.00 | - | |
| .................................................................................................................tgcGTAAGCTCATTGTGGGCAGGGACGT................................................................................................................................................................................................................. | 28 | 1.00 | 0.00 | - | 1.00 | - |