| (1) HEART |
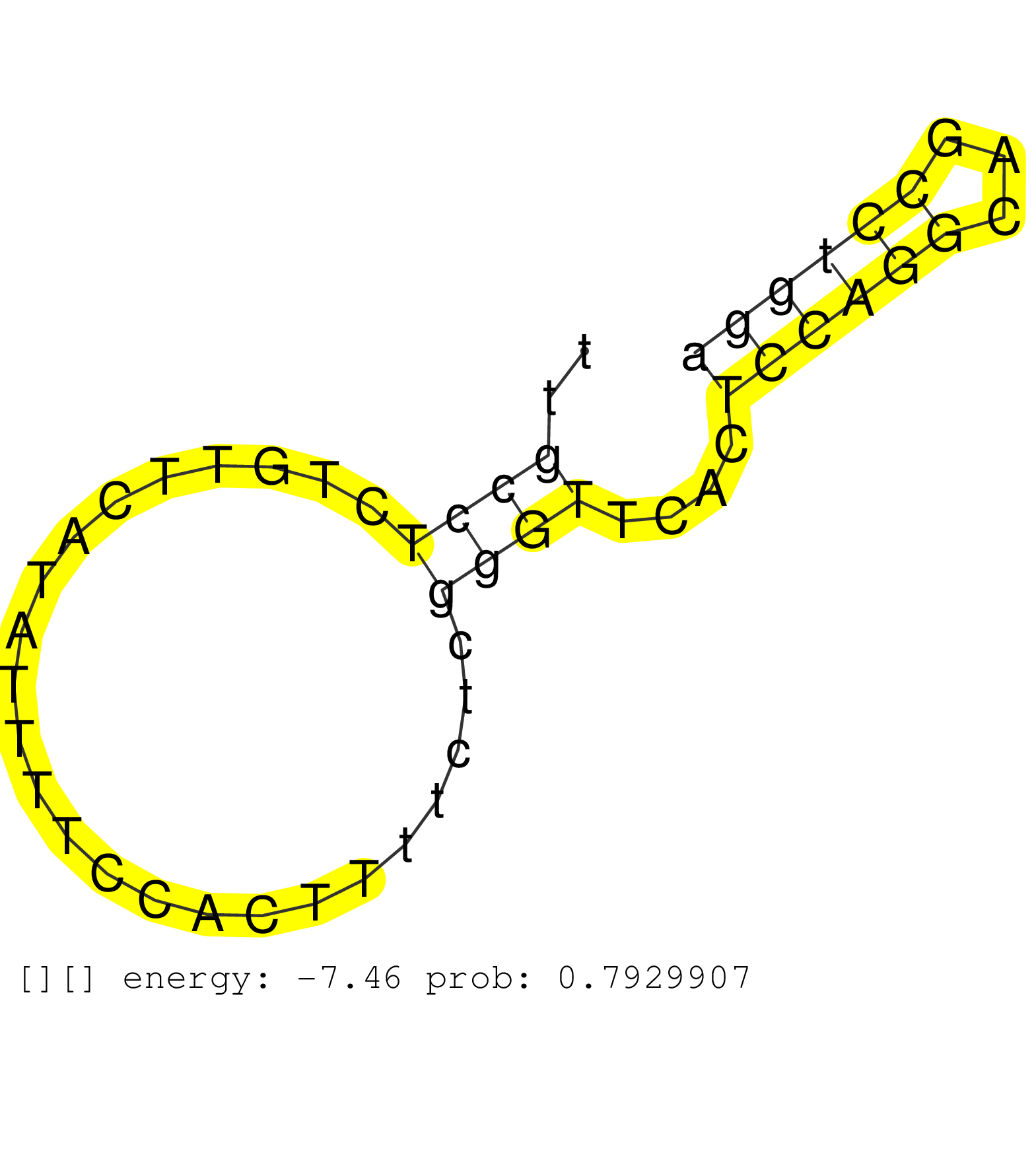
| GTTGACTTATCTCCCTCATGGTTGTCCTAAAACATCTAGTGAAGAGGAAGGTACATGCTGTAGTTGGCCATGATTTAGACCTCAGCTGGTGCTGTGACTGACAGTCACAGGCAATCTGGAAGTCACAAGCAATCTGGTTGGAAGTCACAAGCACACTGTTTCATCTCTTTGCCATTTGTCCAAACTGCAGAGAGCTTTGCCTCTGTTCATATTTTCCACTTTTCTCGGGTTCACTCCAGGCAGCCTGGATACGAGAGGATATATCTCTGGGGTCATTGCAGCAATCTTTTCTCTTTTGGGACAAGGTGAAGGGTACAGGCCTAAGCATTCTAAAGGTTCAGGCTGGCCAG ......................................................................................................................................................................................................((((........................))))....((((((...))))))..................................................................................................... ....................................................................................................................................................................................................197.................................................249................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................................................................................................TGGGACAAGGTGAAGC...................................... | 16 | 6.00 | 0.00 | 6.00 | |
| ..............................................................................................................................................................................TTTGTCCAAACTGCAGAGAGCTTTGC...................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 |
| .....................................AGTGAAGAGGAAGGTACATGCTGTAGTT............................................................................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 |
| ...........................................GAGGAAGGTACATGCTGTAGTTGGCC......................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 |
| .........................................................................................................................................................................................................TCTGTTCATATTTTCCACTT................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | 1.00 |
| ...................................CTAGTGAAGAGGAAGGTACATGCTGTAGT.............................................................................................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 |
| ...........................................GAGGAAGGTACATGCTGTAGTT............................................................................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 |
| ....................................................................................................................................................................................................................................GTTCACTCCAGGCAGCC......................................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 |
| ......................................................................................................................................................................................................................................TCACTCCAGGCAGCCTGGATACGAGAGG............................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 |
| ...................................................................................................................................ATCTGGTTGGAAGTCACAAGCACACT................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 |
| ......................................................................................................................................................................CTTTGCCATTTGTCCAAACTGCA................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | 1.00 |
| ........................................................................................................................................................................................................................................................................................................TGGGACAAGGTGAAGCA..................................... | 17 | 1.00 | 0.00 | 1.00 | |
| .................................................................................................................................CAATCTGGTTGGAAGTGGAC......................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | 1.00 | |
| ..........................................................................................................................................................................................................................................................................................................................................TAAAGGTTCAGGCTGGC... | 17 | 1 | 1.00 | 1.00 | 1.00 |
| ...................................................................................AGCTGGTGCTGTGACT........................................................................................................................................................................................................................................................... | 16 | 2 | 0.50 | 0.50 | 0.50 |
| .......................................................................................................................................................................................................................................CACTCCAGGCAGCCT........................................................................................................ | 15 | 6 | 0.17 | 0.17 | 0.17 |
| GTTGACTTATCTCCCTCATGGTTGTCCTAAAACATCTAGTGAAGAGGAAGGTACATGCTGTAGTTGGCCATGATTTAGACCTCAGCTGGTGCTGTGACTGACAGTCACAGGCAATCTGGAAGTCACAAGCAATCTGGTTGGAAGTCACAAGCACACTGTTTCATCTCTTTGCCATTTGTCCAAACTGCAGAGAGCTTTGCCTCTGTTCATATTTTCCACTTTTCTCGGGTTCACTCCAGGCAGCCTGGATACGAGAGGATATATCTCTGGGGTCATTGCAGCAATCTTTTCTCTTTTGGGACAAGGTGAAGGGTACAGGCCTAAGCATTCTAAAGGTTCAGGCTGGCCAG ......................................................................................................................................................................................................((((........................))))....((((((...))))))..................................................................................................... ....................................................................................................................................................................................................197.................................................249................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) |
|---|