| (1) BRAIN | (1) HEART | (1) KIDNEY | (2) OTHER |
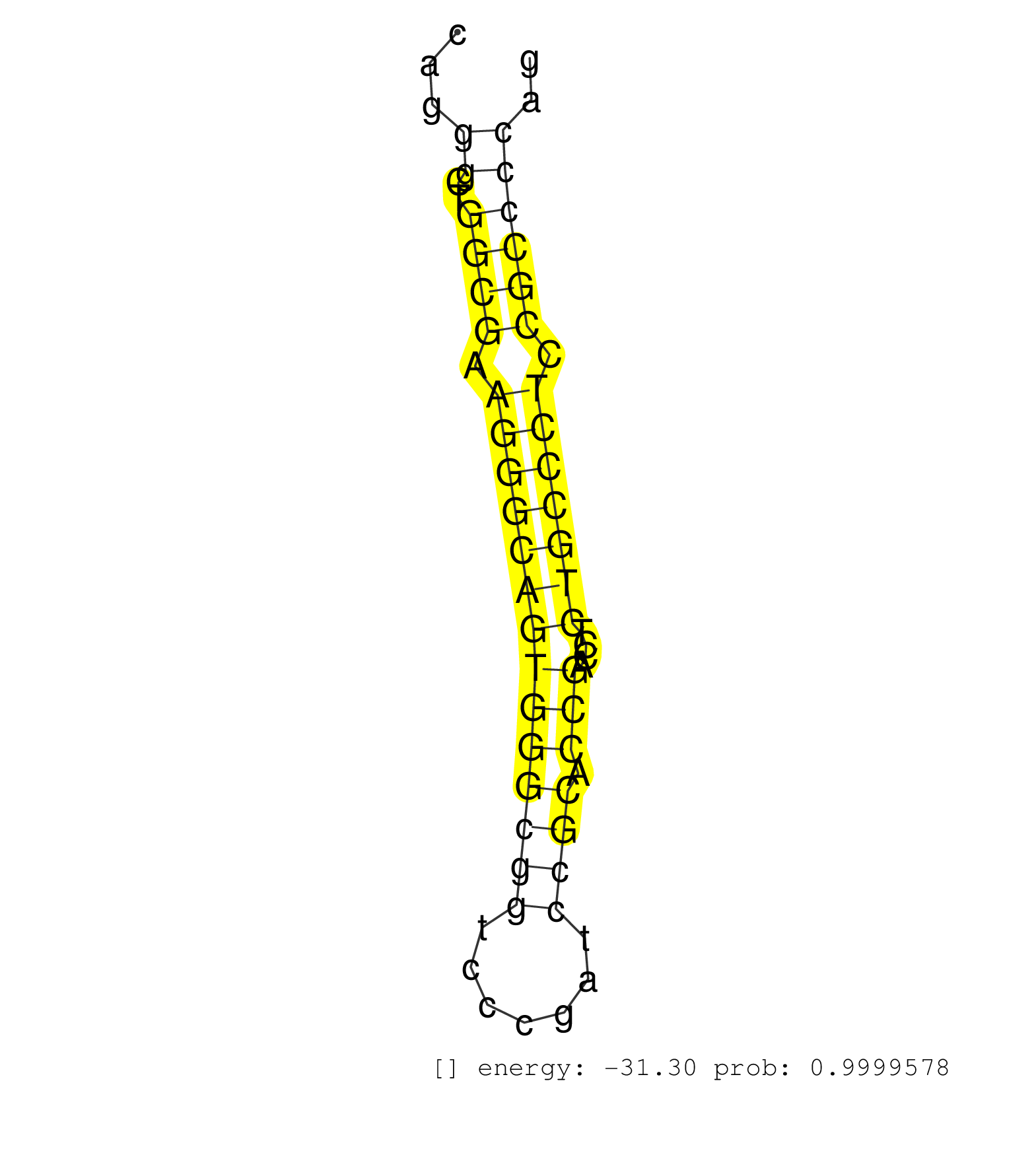
| GCAGGGGGTTTCTGAGGAAGAAGCGGTGGGATCGAGGGATGGATCCACCCCGGGAACCGAACGGTGGGCCGGGGGGAGTCGAGGATGAGTCGAGGTGGCCGCCTTCAGTTACGGGGAGCGTGGCATGGTGTTGCCGAAGGAGGTGGGGACGTCGGGAGGAGGGGAGACTTTTTAGCCATGTTGAGCTTAAGGGCCCTACCCCTAGGGGGGAAGGCAGAGTGAGGAGAGGAAGGGGCTCCCAGGGCTGGCGAAGGGCAGTGGGCGGTCCCGATCCGCACCGACCTCTGCCCTCCGCCCCAGGTACGCCAGCTCCGAGGAGTTCGCTGCCGATGCCCTCCTGGTCTTCGACA ..................................................................................................................................................................................................................................................((..((((.((((((((((((((.......)))).)))....))))))).)))))).................................................... ...............................................................................................................................................................................................................................................240.........................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553592(SRX182798) source: Brain. (Brain) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................................................CTGGCGAAGGGCAGTGGG........................................................................................ | 18 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - |
| ...................................................................................................................................................................................................................................................GCTGGCGAAGGGCAGTGGGCGGTA................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| ...................................................................................................................................................................................................................................................GCTGGCGAAGGGCAGTGGGCGGA.................................................................................... | 23 | 1.00 | 0.00 | - | 1.00 | - | - | - | |
| ....................................................................................................................................................................................................................................................CTGGCGAAGGGCAGTGGGC....................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................................................................................CTGGCGAAGGGCAGTGGGCGGTT................................................................................... | 23 | 1.00 | 1.00 | - | 1.00 | - | - | - | |
| ....................................................................................................................................................................................................................................................CTGGCGAAGGGCAGTGGGCGGT.................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................................................................................CTGGCGAAGGGCAGTGGGCGGTTT.................................................................................. | 24 | 1.00 | 1.00 | - | 1.00 | - | - | - | |
| .....................................................................................................................................................................................................................................................................................................................................GCCGATGCCCTCCTGGTA....... | 18 | 1.00 | 0.00 | - | - | 1.00 | - | - | |
| ....................................................................................................................................................................................................................................................CTGGCGAAGGGCAGTGGGCGGTAAA................................................................................. | 25 | 1.00 | 1.00 | - | 1.00 | - | - | - | |
| ..................................................................................................................................................................................................................................................................................GCACCGACCTCTGCCCTCCGC....................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................................................................................TGGCGAAGGGCAGTGGGCGGTCATTT............................................................................... | 26 | 1.00 | 0.00 | - | - | 1.00 | - | - | |
| ...................................................................................................................................................................................................................................................GCTGGCGAAGGGCAGTGGGCGAA.................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | 1.00 | |
| ....................................................................................................................................................................................................................................................CTGGCGAAGGGCAGTGG......................................................................................... | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................................................................................CTGGCGAAGGGCAGTGGGCGGTA................................................................................... | 23 | 1.00 | 1.00 | - | 1.00 | - | - | - | |
| .....................................................................................................................................................................................................................................................TGGCGAAGGGCAGTGGGCGT..................................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - |
| GCAGGGGGTTTCTGAGGAAGAAGCGGTGGGATCGAGGGATGGATCCACCCCGGGAACCGAACGGTGGGCCGGGGGGAGTCGAGGATGAGTCGAGGTGGCCGCCTTCAGTTACGGGGAGCGTGGCATGGTGTTGCCGAAGGAGGTGGGGACGTCGGGAGGAGGGGAGACTTTTTAGCCATGTTGAGCTTAAGGGCCCTACCCCTAGGGGGGAAGGCAGAGTGAGGAGAGGAAGGGGCTCCCAGGGCTGGCGAAGGGCAGTGGGCGGTCCCGATCCGCACCGACCTCTGCCCTCCGCCCCAGGTACGCCAGCTCCGAGGAGTTCGCTGCCGATGCCCTCCTGGTCTTCGACA ..................................................................................................................................................................................................................................................((..((((.((((((((((((((.......)))).)))....))))))).)))))).................................................... ...............................................................................................................................................................................................................................................240.........................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553592(SRX182798) source: Brain. (Brain) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|