| (1) HEART | (1) KIDNEY | (2) OTHER |
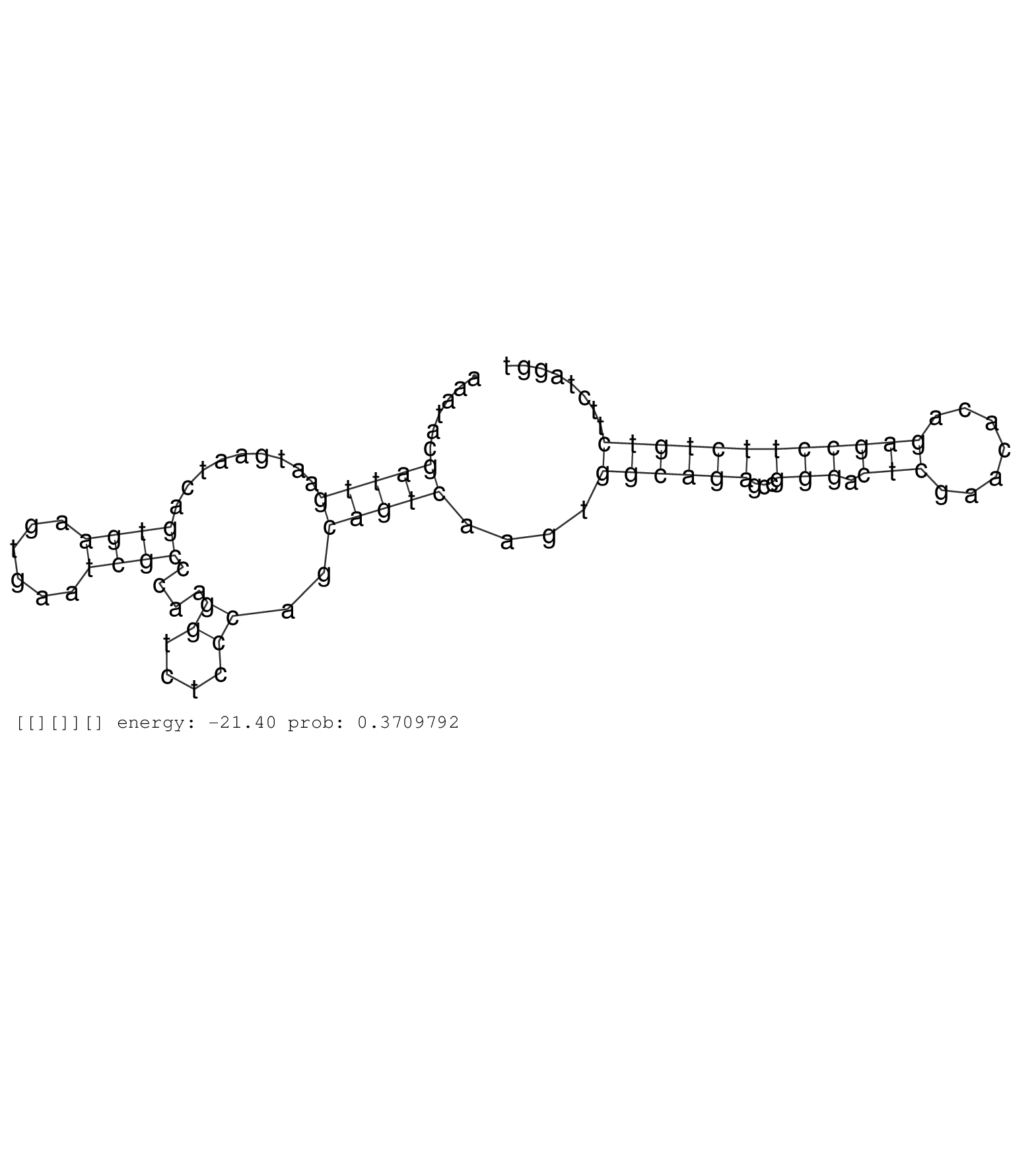
| CTTCACTTCTCGGTGCTTCAGTTGCCTCATCTGTAAAATGGGGATTAAGACTGTGAGCCCCACATGGGACACCCTGATTATCTTGGATCTACTCCAGCGCTTAGGACGGTGCTCGGCACATAGTGAGCACTTAACAAATACCATAATTATTATTAATTAGTAGTAGTAGTGCAGTGCTTTGCTCCCAGCAAGCACTCAATAAATACGATTGAATGAATCAGTGAAGTGAATCGCCCAAGGTCTCCCAGCAGTCAAGTGGCAGAGCCGGGACTCGAACACAGAGCCTTCTGTCTTCTAGGTGAAATCGGTGCTGGTGCCGCCAATGCCCGTGAAGACAGAGAAGGCCCAAT ..............................................................................................................................................................................................................(((((.........((((......))))....((....))..)))))....((((((...(((.(((.......)))))))))))).......................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|---|
| ........................................................................................................GACGGTGCTCGGCACATAGA.................................................................................................................................................................................................................................. | 20 | 5.00 | 0.00 | 2.00 | 2.00 | 1.00 | - | |
| ........................................................................................................GACGGTGCTCGGCACATAGTAGA............................................................................................................................................................................................................................... | 23 | 2.00 | 0.00 | - | 2.00 | - | - | |
| ......................................................................................................AGGACGGTGCTCGGCACATAGA.................................................................................................................................................................................................................................. | 22 | 2.00 | 0.00 | 2.00 | - | - | - | |
| ......................................................................................................AGGACGGTGCTCGGCACATAGTT................................................................................................................................................................................................................................. | 23 | 2.00 | 0.00 | 1.00 | - | 1.00 | - | |
| ......................................................................................................AGGACGGTGCTCGGCACATAGTTTAT.............................................................................................................................................................................................................................. | 26 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ......................................................................................................AGGACGGTGCTCGGCACATAGAAT................................................................................................................................................................................................................................ | 24 | 1.00 | 0.00 | - | - | 1.00 | - | |
| ............................................................................................................................................................................................................................................................................................................................CCGCCAATGCCCGTGAAGACA............. | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ......................................................................................................AGGACGGTGCTCGGCACATAGAT................................................................................................................................................................................................................................. | 23 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ...........................................................................................................................................................................................................TACGATTGAATGAATCAGTGAAGGAAT........................................................................................................................ | 27 | 1.00 | 0.00 | - | - | - | 1.00 | |
| .........................................GGATTAAGACTGTGAGCCCCACATGTGC......................................................................................................................................................................................................................................................................................... | 28 | 1.00 | 0.00 | - | - | 1.00 | - | |
| ........................................................................................................GACGGTGCTCGGCACATAGAA................................................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ........................................................................................................GACGGTGCTCGGCACATAGAACA............................................................................................................................................................................................................................... | 23 | 1.00 | 0.00 | 1.00 | - | - | - |
| CTTCACTTCTCGGTGCTTCAGTTGCCTCATCTGTAAAATGGGGATTAAGACTGTGAGCCCCACATGGGACACCCTGATTATCTTGGATCTACTCCAGCGCTTAGGACGGTGCTCGGCACATAGTGAGCACTTAACAAATACCATAATTATTATTAATTAGTAGTAGTAGTGCAGTGCTTTGCTCCCAGCAAGCACTCAATAAATACGATTGAATGAATCAGTGAAGTGAATCGCCCAAGGTCTCCCAGCAGTCAAGTGGCAGAGCCGGGACTCGAACACAGAGCCTTCTGTCTTCTAGGTGAAATCGGTGCTGGTGCCGCCAATGCCCGTGAAGACAGAGAAGGCCCAAT ..............................................................................................................................................................................................................(((((.........((((......))))....((....))..)))))....((((((...(((.(((.......)))))))))))).......................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................................................................................................................................gcaTGGGCCTTCTCTACG | 18 | 1.00 | 0.00 | 1.00 | - | - | - |