| (1) BRAIN | (1) HEART | (1) KIDNEY | (1) OTHER |
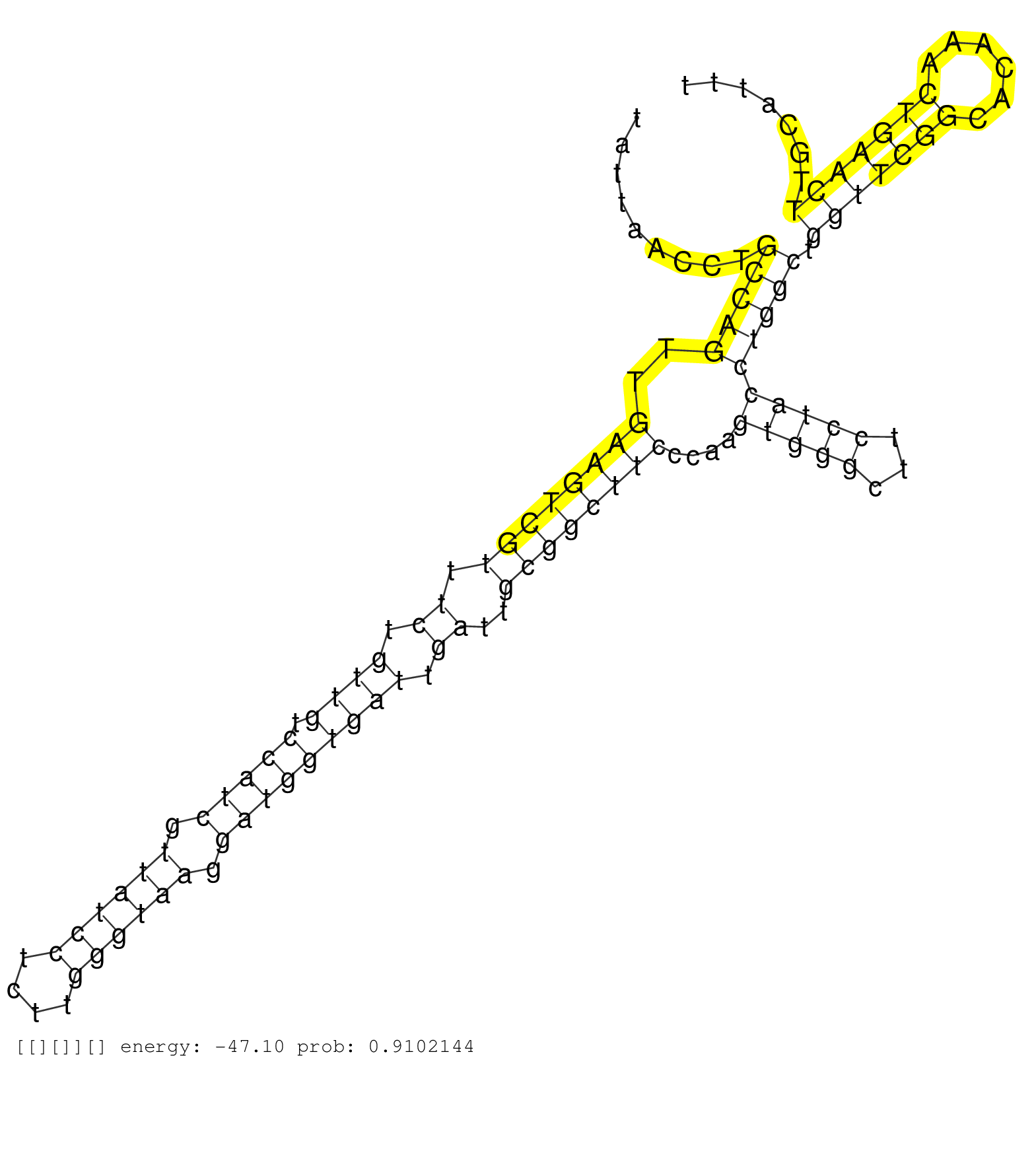
| GGAACCTCTACCTGAGGACTATTACACGAGACCAGTAAATTTAACAGAGGGTAAAATTGCCTTTTCATTTCCCTTCATGTCCAGTGCTGTTTTTTATCTATCATTAACAAAAAAACCAGCTTTAGCCCCTTTAAATCCTAGTTTCCTATAAAATGAAGCACATTATGAGCTAAGTCTTTCCTCCTGAAACAGCAGTGCAGTATCTTAAAGAGTTTTGACTTTTGAAAGGTTAAAGGGGTGTCAAATGGGTATTAACCTGCCAGTTGAAGTCGTTTCTGTTGTCCATCGTTATCCTCTTGGGTAAGGATGGTGATTGATTGCGGCTTCCCAAGTGGGCTTCCTACCTGGCTGGTTCGGCACAAACTGAACTTGCATTTCTTGTCTGTGTGCTAGTGACCACCCTACGTCAGCGTCTCCTGCAGCCCGATTTCCAACCAGTCTGC ..................................................................................................................................................................................................................................................................(((((..((((((((.((.((((.(((((.((((((....)))))).))))))))).))..))))))))....(((((...)))))))))).(((((((......)))))))......................................................................... .........................................................................................................................................................................................................................................................250............................................................................................................................377................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553592(SRX182798) source: Brain. (Brain) |
|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................................................ACCTGCCAGTTGAAGAAC........................................................................................................................................................................... | 18 | 4.00 | 0.00 | 4.00 | - | - | - | |
| ..............................................................................................................................................................................................................................................................ACCTGCCAGTTGAAGAACT.......................................................................................................................................................................... | 19 | 3.00 | 0.00 | 1.00 | - | 2.00 | - | |
| .............................................................................................................................................................................................................................................................AACCTGCCAGTTGAAGAACT.......................................................................................................................................................................... | 20 | 3.00 | 0.00 | 1.00 | 2.00 | - | - | |
| .....................TTACACGAGACCAGTAAATTTAACAGAGGTGA...................................................................................................................................................................................................................................................................................................................................................................................................... | 32 | 2.00 | 0.00 | - | 2.00 | - | - | |
| ..............................................................................................................................................................................................................................................................ACCTGCCAGTTGAAGAAAC.......................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | 1.00 | |
| ..............AGGACTATTACACGAGACCAGTAAATTTAACA............................................................................................................................................................................................................................................................................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .................................................................................................................................................................................................................................................................................................................................................................TCGGCACAAACTGAACTTGC...................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ......................TACACGAGACCAGTAAATTTAACAGAGGT........................................................................................................................................................................................................................................................................................................................................................................................................ | 29 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .....................TTACACGAGACCAGTAAATTTAACAGAG.......................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................CAGCGTCTCCTGCAGC.................... | 16 | 2 | 0.50 | 0.50 | - | - | 0.50 | - |
| ........TACCTGAGGACTATTACACGAGAC........................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 2 | 0.50 | 0.50 | 0.50 | - | - | - |
| ..............AGGACTATTACACGAGACC.......................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | 0.50 | - |
| GGAACCTCTACCTGAGGACTATTACACGAGACCAGTAAATTTAACAGAGGGTAAAATTGCCTTTTCATTTCCCTTCATGTCCAGTGCTGTTTTTTATCTATCATTAACAAAAAAACCAGCTTTAGCCCCTTTAAATCCTAGTTTCCTATAAAATGAAGCACATTATGAGCTAAGTCTTTCCTCCTGAAACAGCAGTGCAGTATCTTAAAGAGTTTTGACTTTTGAAAGGTTAAAGGGGTGTCAAATGGGTATTAACCTGCCAGTTGAAGTCGTTTCTGTTGTCCATCGTTATCCTCTTGGGTAAGGATGGTGATTGATTGCGGCTTCCCAAGTGGGCTTCCTACCTGGCTGGTTCGGCACAAACTGAACTTGCATTTCTTGTCTGTGTGCTAGTGACCACCCTACGTCAGCGTCTCCTGCAGCCCGATTTCCAACCAGTCTGC ..................................................................................................................................................................................................................................................................(((((..((((((((.((.((((.(((((.((((((....)))))).))))))))).))..))))))))....(((((...)))))))))).(((((((......)))))))......................................................................... .........................................................................................................................................................................................................................................................250............................................................................................................................377................................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553592(SRX182798) source: Brain. (Brain) |
|---|