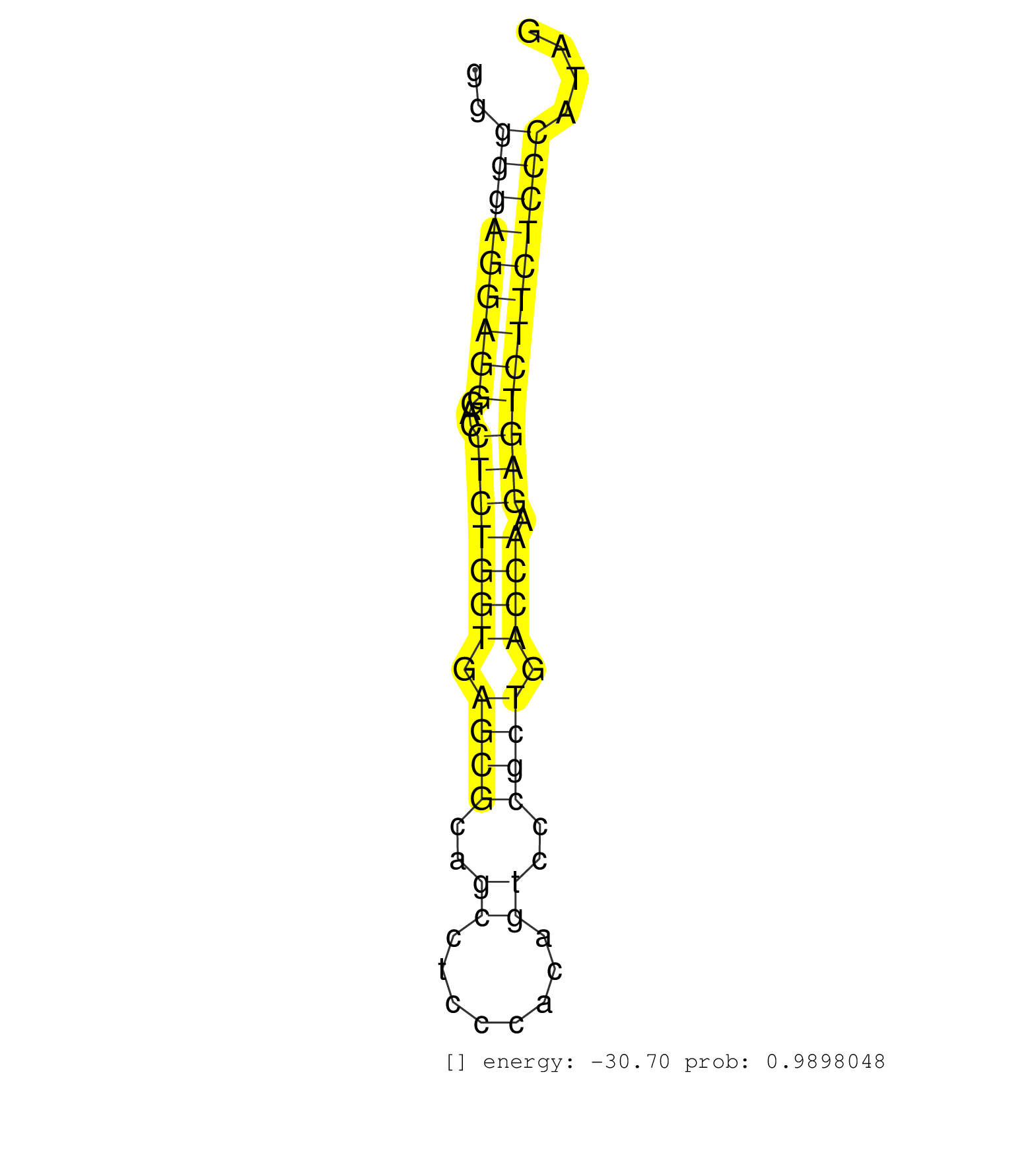
| (1) AGO.mut | (4) AGO2.ip | (2) BRAIN | (2) CELL-LINE | (1) DCR.mut | (5) EMBRYO | (7) FIBROBLAST | (1) KIDNEY | (14) LIVER | (11) OTHER | (9) OTHER.mut | (1) PIWI.ip | (1) PIWI.mut | (1) SKIN | (1) SPLEEN | (16) TESTES | (1) UTERUS |
| GCTTCTCTGTGCAGCGCTGCTGCTGCTGGCCACCAGCCGCCTGGCCACAGGTAGGTCTCGCCACTGCCACTGGGGGAGGAGGGACCTCTGGTGAGCGCAGCCTCCCACAGTCCCGCTGACCAAGAGTCTTCTCCCATAGGGGCGCCTATCGCCAATGAGCTGCGCTGTCAGTGCCTGCAGACCATGGCT .........................................................................(((((((((...(((((((.((((..((........))..)))).)))).))))))))))))...................................................... .......................................................................72.................................................................139................................................ | Size | Perfect hit | Total Norm | Perfect Norm | mjLiverKO2() Liver Data. (Zcchc11 liver) | mjTestesWT1() Testes Data. (testes) | mjLiverWT1() Liver Data. (liver) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | mjLiverWT3() Liver Data. (liver) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR059775(GSM562837) MEF_Drosha. (MEF) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR685340(GSM1079784) "Small RNAs (15-50 nts in length) from immort. (dicer cell line) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR029123(GSM416611) NIH3T3. (cell line) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR065048(SRR065048) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR038744(GSM527279) small RNA-Seq. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................AGGAGGGACCTCTGGTGAGCG............................................................................................ | 21 | 1 | 179.00 | 179.00 | 77.00 | 32.00 | 17.00 | 3.00 | 4.00 | 4.00 | 11.00 | 3.00 | 4.00 | 2.00 | 10.00 | - | - | - | - | - | 4.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCGA........................................................................................... | 22 | 1 | 97.00 | 179.00 | 46.00 | - | 3.00 | 12.00 | 6.00 | - | 4.00 | 2.00 | 8.00 | 5.00 | - | 1.00 | - | - | 1.00 | 2.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GAGGAGGGACCTCTGGTGAGCG............................................................................................ | 22 | 1 | 68.00 | 68.00 | 49.00 | 11.00 | - | 2.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCGT........................................................................................... | 22 | 1 | 52.00 | 179.00 | 10.00 | - | 12.00 | 3.00 | 3.00 | 8.00 | 3.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGC............................................................................................. | 20 | 1 | 29.00 | 29.00 | - | - | 6.00 | 2.00 | 6.00 | - | - | 8.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCGAAT......................................................................................... | 24 | 1 | 6.00 | 179.00 | - | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GAGGAGGGACCTCTGGTGAGCGT........................................................................................... | 23 | 1 | 6.00 | 68.00 | - | - | - | - | 2.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGGGGGAGGAGGGACCTCTGGTGAGCG............................................................................................ | 27 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCGAA.......................................................................................... | 23 | 1 | 3.00 | 179.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................GGAGGGACCTCTGGTGAGCG............................................................................................ | 20 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GAGGAGGGACCTCTGGTGAGCGAAA......................................................................................... | 25 | 1 | 3.00 | 68.00 | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GAGGAGGGACCTCTGGTGAGCGATT......................................................................................... | 25 | 1 | 3.00 | 68.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCGAAA......................................................................................... | 24 | 1 | 3.00 | 179.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................GGAGGGACCTCTGGTGAGCGT........................................................................................... | 21 | 1 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTCTCGCCACTGCCACTGGGGGAGGAGGGACCTCTGGTGAGCGCAGCCTCCCACAGTCCCGCTGACCAAGAGTCTTCTCCCATAG.................................................. | 89 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCGAT.......................................................................................... | 23 | 1 | 2.00 | 179.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................ATGAGCTGCGCTGTCAGTGCC.............. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................GGAGGGACCTCTGGTGAGCGA........................................................................................... | 21 | 1 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCGCAC......................................................................................... | 24 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................GAGGAGGGACCTCTGGTGAGC............................................................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TTCTCTGTGCAGCGCTGCTGCTGCTGGCC.............................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGAG............................................................................................ | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................GGAGGGACCTCTGGTGAGC............................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAG.............................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GAGGAGGGACCTCTGGTGAGCGAAGA........................................................................................ | 26 | 1 | 1.00 | 68.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................GGAGGGACCTCTGGTGAG.............................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCGC........................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................CGCCACTGCCACTGGGGGA................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCGTT.......................................................................................... | 23 | 1 | 1.00 | 179.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCGG........................................................................................... | 22 | 1 | 1.00 | 179.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GAGGAGGGACCTCTGGTGAGTG............................................................................................ | 22 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................AGGAGGGACCTCTGGTGAGCGAGA......................................................................................... | 24 | 1 | 1.00 | 179.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................CCAATGAGCTGCGCTGTC.................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GAGGAGGGACCTCTGGTGAGCGA........................................................................................... | 23 | 1 | 1.00 | 68.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGGGCG............................................................................................ | 21 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................AGGAGGGACCTCTGGTGAGCGTGA......................................................................................... | 24 | 1 | 1.00 | 179.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGAGA........................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTCTCGCCACTGCCACTGGGGGAGGAGGGACCTCTGGTGAGCGCAGCCTCCCACAGTCCCGCTGACCAAGAGTCTTCTCCCATAGA................................................. | 90 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCAT........................................................................................... | 22 | 1 | 1.00 | 29.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCGTAAA........................................................................................ | 25 | 1 | 1.00 | 179.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCGAG.......................................................................................... | 23 | 1 | 1.00 | 179.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGAGGGACCTCTGGTGAGCGAAGT........................................................................................ | 25 | 1 | 1.00 | 179.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GAGGAGGGACCTCTGGTGAGCGAAT......................................................................................... | 25 | 1 | 1.00 | 68.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGACCAAGAGTCTTCTCCCATTT.................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................AGGAGGGACCTCTGGTGAGCGATAA........................................................................................ | 25 | 1 | 1.00 | 179.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| GCTTCTCTGTGCAGCGCTGCTGCTGCTGGCCACCAGCCGCCTGGCCACAGGTAGGTCTCGCCACTGCCACTGGGGGAGGAGGGACCTCTGGTGAGCGCAGCCTCCCACAGTCCCGCTGACCAAGAGTCTTCTCCCATAGGGGCGCCTATCGCCAATGAGCTGCGCTGTCAGTGCCTGCAGACCATGGCT .........................................................................(((((((((...(((((((.((((..((........))..)))).)))).))))))))))))...................................................... .......................................................................72.................................................................139................................................ | Size | Perfect hit | Total Norm | Perfect Norm | mjLiverKO2() Liver Data. (Zcchc11 liver) | mjTestesWT1() Testes Data. (testes) | mjLiverWT1() Liver Data. (liver) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | mjLiverWT3() Liver Data. (liver) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR059775(GSM562837) MEF_Drosha. (MEF) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR685340(GSM1079784) "Small RNAs (15-50 nts in length) from immort. (dicer cell line) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR029123(GSM416611) NIH3T3. (cell line) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR065048(SRR065048) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR038744(GSM527279) small RNA-Seq. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................GCTGCGCTGTCAGTGCCTGCAGACCA..... | 26 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................CCGCTGACCAAGAGTCTTCTCCCA..................................................... | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TCCCGCTGACCAAGAGTCTTCTCCCA..................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TCCCACAGTCCCGCTA....................................................................... | 16 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................ACAGTCCCGCTGACCAAGAGTCTTC.......................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................CCAGCCGCCTGGCCACAGGTCG....................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................GCCTCCCACAGTCCCGCTGACCAAGA................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................CTCCCACAGTCCCGCTGACCAATA................................................................ | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................CGCCTGGCCACAGGTAGG...................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................TCGCCACTGCCACTGGGG.................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................GCCACAGGTAGGTCTCGCCACTGCCA........................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GCCTCCCACAGTCCCGCTGACCAA.................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................GGCCACCAGCCGCCTCGG................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................GGGGAGGAGGGACCTCTGGTGAGCGCA.......................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................CACAGTCCCGCTGACCAAGAGTCTT........................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CAGGTAGGTCTCGCCACT............................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............CGCTGCTGCTGCTGGCCACCAGCCG...................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................CCAGCCGCCTGGCCACAGGTAG....................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................CCTCCCACAGTCCCGCTGACCAA.................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GTCCCGCTGACCAAGAGTCTTCTCCCA..................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................CTGCGCTGTCAGTGCCTGCAGACCA..... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................GCCACAGGTAGGTCTCGCCA.............................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................CCGCTGACCAAGAGTCTTCTCCCATA................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................GCCTGGCCACAGGTAGGT..................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................GCTGCTGCTGGCCACCA.......................................................................................................................................................... | 17 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |