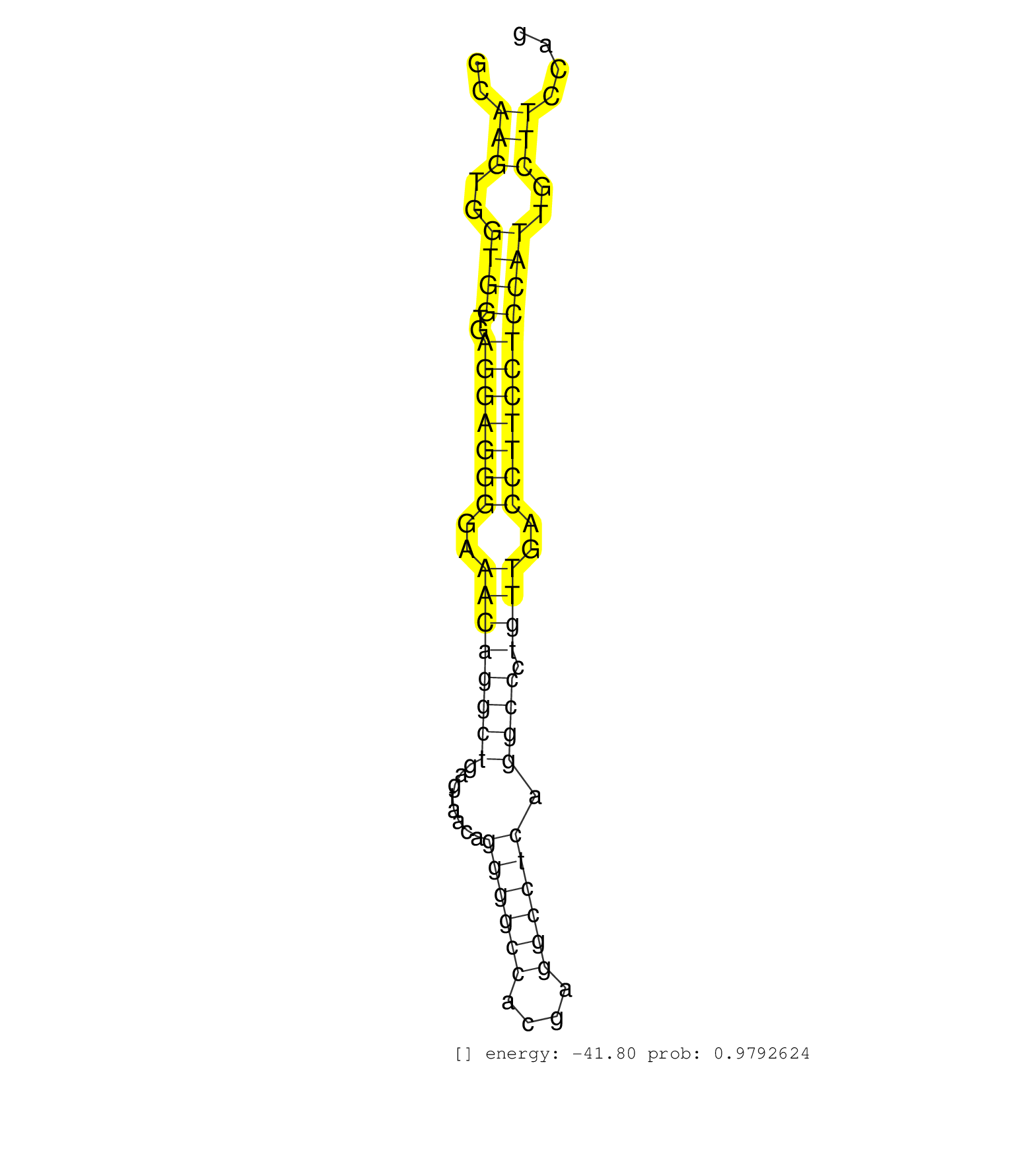
| (1) AGO.mut | (2) AGO1.ip | (2) AGO2.ip | (1) AGO3.ip | (1) BRAIN | (1) CELL-LINE | (1) EMBRYO | (2) ESC | (1) KIDNEY | (7) LIVER | (4) OTHER | (4) OTHER.mut | (2) PIWI.ip | (4) SKIN | (5) TESTES | (5) TOTAL-RNA |
| GCACACACAGCTATACCACTCGAGTCTGTGCCATCTCTTCCAGAAGAAAGGCAAGTGGTGGTGAGGAGGGGAAACAGGCTGAGTAACAGGGGCCACGAGGCCTCAGGCCCTGTTGACCTTCCTCCATTGCTTCCAGATTATGTGCGGACCTGCATTATGGACAATGGGGTCAGCTACCGGGGCACT ....................................................(((..((((..(((((((..((((((((........((((((....)))))).)))).))))..)))))))))))..)))...................................................... ..................................................51...................................................................................136................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | mjLiverKO3() Liver Data. (Zcchc11 liver) | mjLiverKO1() Liver Data. (Zcchc11 liver) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM510456(GSM510456) newborn_rep12. (total RNA) | mjLiverWT3() Liver Data. (liver) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR037914(GSM510451) newborn_rep7. (total RNA) | GSM510450(GSM510450) newborn_rep6. (total RNA) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | GSM640583(GSM640583) small RNA in the liver with paternal Low prot. (liver) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR073955(GSM629281) total RNA. (blood) | SRR037937(GSM510475) 293cand2. (cell line) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................TTGACCTTCCTCCATTGCTTCC.................................................... | 22 | 1 | 77.00 | 77.00 | 48.00 | 17.00 | 3.00 | - | 1.00 | 4.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCCA................................................... | 23 | 1 | 21.00 | 21.00 | 8.00 | 3.00 | 9.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCCAT.................................................. | 24 | 1 | 16.00 | 21.00 | 6.00 | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTC..................................................... | 21 | 1 | 15.00 | 15.00 | 9.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCA.................................................... | 22 | 1 | 12.00 | 15.00 | 2.00 | 1.00 | 3.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCCT................................................... | 23 | 1 | 9.00 | 77.00 | 6.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCAAGTGGTGGTGAGGAGGGGAAAC............................................................................................................... | 25 | 1 | 8.00 | 8.00 | 3.00 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCT.................................................... | 22 | 1 | 7.00 | 15.00 | 4.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTT...................................................... | 20 | 1 | 6.00 | 6.00 | 3.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCAAGTGGTGGTGAGGAGGGGA.................................................................................................................. | 22 | 1 | 4.00 | 4.00 | - | - | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCCAGT................................................. | 25 | 4.00 | 0.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ..................................................GCAAGTGGTGGTGAGGAGGGGAAT................................................................................................................ | 24 | 1 | 4.00 | 2.00 | - | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................CAAGTGGTGGTGAGGAGGGGAAT................................................................................................................ | 23 | 3.00 | 0.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................TATGGACAATGGGGTATT............. | 18 | 3.00 | 0.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................TTGACCTTCCTCCATTGCTTA..................................................... | 21 | 1 | 3.00 | 6.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCT....................................................... | 19 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................ACTCGAGTCTGTGCCATCTCTTCCAGAAGA........................................................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCCAA.................................................. | 24 | 1 | 2.00 | 21.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTAA.................................................... | 22 | 1 | 2.00 | 6.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCAAGTGGTGGTGAGGAGGGGAAATTT............................................................................................................. | 27 | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GCAAGTGGTGGTGAGGAGGGGAA................................................................................................................. | 23 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCAAGTGGTGGTGAGGAGGGGAAACT.............................................................................................................. | 26 | 1 | 1.00 | 8.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCAAT.................................................. | 24 | 1 | 1.00 | 15.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCCAGTATT.............................................. | 28 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCCAGTTT............................................... | 27 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................GACCTTCCTCCATTGCTTCCAGA................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCCC................................................... | 23 | 1 | 1.00 | 77.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCCATAAT............................................... | 27 | 1 | 1.00 | 21.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCAAGTGGTGGTGAGGAGGGGAT................................................................................................................. | 23 | 1 | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................GGGAAACAGGCTGAGTAAC................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCCTTA................................................. | 25 | 1 | 1.00 | 77.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................CATTATGGACAATGGGG................. | 17 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCCG................................................... | 23 | 1 | 1.00 | 77.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................CACTCGAGTCTGTGCCATCTCTTCCA................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCAAGTGGTGGTGAGGAGGGTAAT................................................................................................................ | 24 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...CACACAGCTATACCACTCG.................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GCAAGTGGTGGTGAGGAGGGAAA................................................................................................................. | 23 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................GGCCACGAGGCCTCATGGA............................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ..................................................GCAAGTGGTGGTGAGGAG...................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TTATGGACAATGGGGTCAGCTACCGG...... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................ATGGGGTCAGCTACCGG...... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GTGCGGACCTGCATTATGGACAATGGGG................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCCTT.................................................. | 24 | 1 | 1.00 | 77.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTGACCTTCCTCCATTGCTTCCACC................................................. | 25 | 1 | 1.00 | 21.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................AGGGGAAACAGGCTGTC....................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ................................................................................................................TTGACCTTCCTCCATTGC........................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGGTGGTGAGGAGGG.................................................................................................................... | 16 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| GCACACACAGCTATACCACTCGAGTCTGTGCCATCTCTTCCAGAAGAAAGGCAAGTGGTGGTGAGGAGGGGAAACAGGCTGAGTAACAGGGGCCACGAGGCCTCAGGCCCTGTTGACCTTCCTCCATTGCTTCCAGATTATGTGCGGACCTGCATTATGGACAATGGGGTCAGCTACCGGGGCACT ....................................................(((..((((..(((((((..((((((((........((((((....)))))).)))).))))..)))))))))))..)))...................................................... ..................................................51...................................................................................136................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | mjLiverKO3() Liver Data. (Zcchc11 liver) | mjLiverKO1() Liver Data. (Zcchc11 liver) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM510456(GSM510456) newborn_rep12. (total RNA) | mjLiverWT3() Liver Data. (liver) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR037914(GSM510451) newborn_rep7. (total RNA) | GSM510450(GSM510450) newborn_rep6. (total RNA) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | GSM640583(GSM640583) small RNA in the liver with paternal Low prot. (liver) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR073955(GSM629281) total RNA. (blood) | SRR037937(GSM510475) 293cand2. (cell line) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................TGTGCCATCTCTTCCAGAACTCG......................................................................................................................................... | 23 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................GACCTTCCTCCATTGCTTCCAGAT................................................ | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................CACTCGAGTCTGTGCCATCTCTTCCAT............................................................................................................................................... | 27 | 1 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................ACTCGAGTCTGTGCCATCTCTTCCAGAA............................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................CAATGGGGTCAGCTAAT........ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .....................GAGTCTGTGCCATCTCTTCCAGAAGAAA......................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................CCTGTTGACCTTCCTCATGC.......................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |