| (3) AGO1.ip | (4) AGO2.ip | (9) B-CELL | (1) BRAIN | (4) EMBRYO | (2) LIVER | (1) LUNG | (5) LYMPH | (19) OTHER | (1) OTHER.mut | (9) SPLEEN | (1) TESTES | (4) THYMUS | (1) TOTAL-RNA |
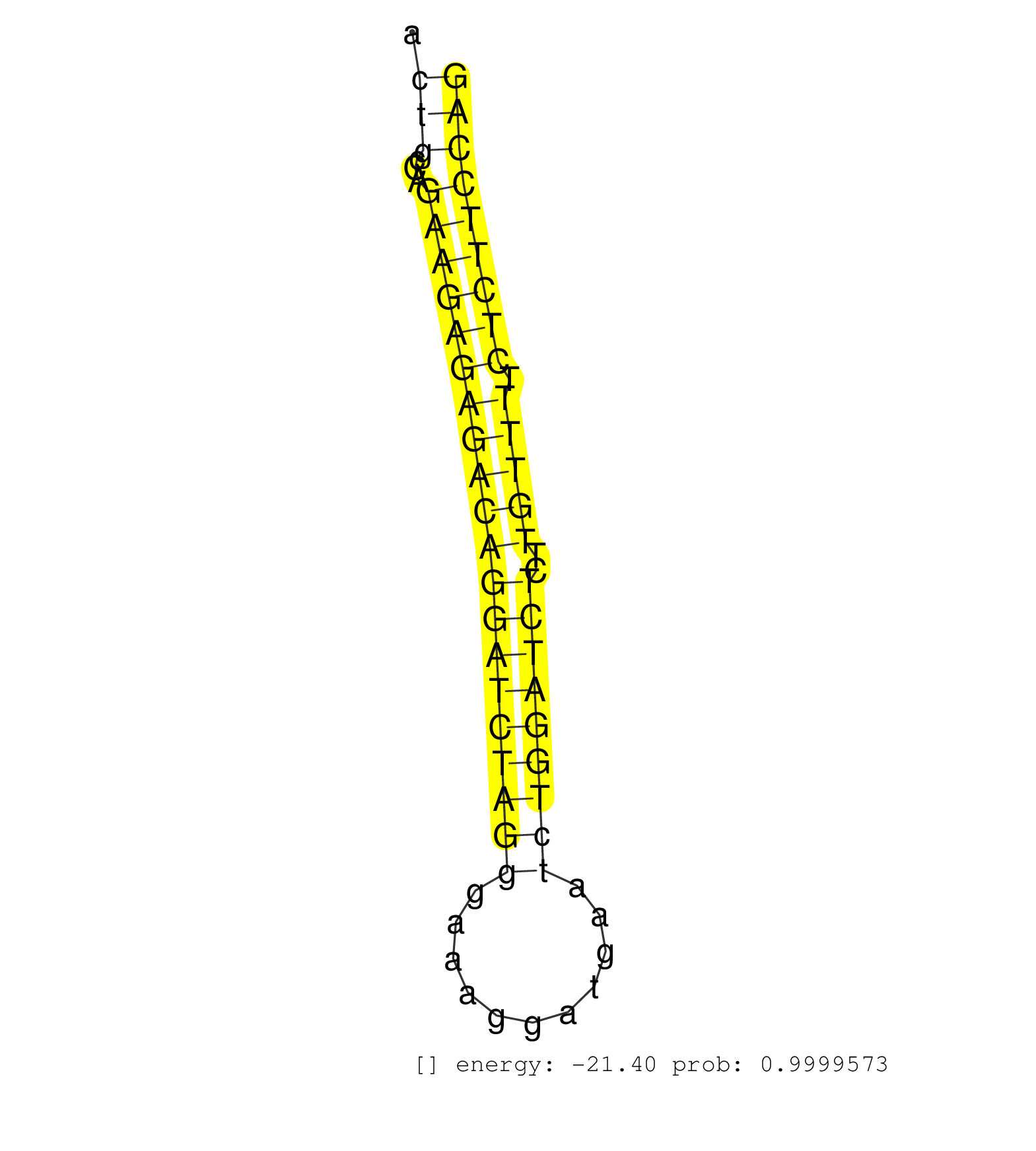
| GGAAACGGATAGATTGTCAACCAGAGACCTGAACTCCTGAGCTAGGAAAGACTTGAGTATTCGCAGGCCACTTTTGATAACGTGGAGGTCTAAACCTTCGATTTTCTTTAAGCCCTACCACCACCACCACACAGTTACTGCCAGAAGAGAGACAGGATCTAGGGAAAGGATGAATCTGGATCTCTTGTTTTCTCTTCCAGGGTCACCGGGGCAACAGCCTGGACAGGAGGTCCCAGGGCGGCCCTCACCC .........................................................................................................................................(((...((((((((((((((((((((...........)))))))))..))))).))))))))).................................................. ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR059768(GSM562830) Treg_control. (spleen) | mjLiverWT1() Liver Data. (liver) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR059765(GSM562827) DN3_control. (thymus) | SRR525243(SRA056111/SRX170319) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR525244(SRA056111/SRX170320) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR525240(SRA056111/SRX170316) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR525241(SRA056111/SRX170317) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR525238(SRA056111/SRX170314) Second IgG control. (Blood) | SRR073954(GSM629280) total RNA. (blood) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR073955(GSM629281) total RNA. (blood) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR525242(SRA056111/SRX170318) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR525239(SRA056111/SRX170315) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR065056(SRR065056) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAG........................................................................................ | 21 | 1 | 49.00 | 49.00 | 13.00 | 7.00 | 3.00 | 7.00 | 4.00 | 1.00 | - | 3.00 | 1.00 | - | - | - | - | 2.00 | - | 2.00 | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAGT....................................................................................... | 22 | 1 | 35.00 | 49.00 | 10.00 | 1.00 | 2.00 | 1.00 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | 1.00 | 2.00 | 2.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGG....................................................................................... | 21 | 1 | 20.00 | 20.00 | 5.00 | - | 3.00 | 1.00 | - | 2.00 | - | - | - | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAG........................................................................................ | 20 | 1 | 18.00 | 18.00 | 3.00 | 2.00 | 1.00 | - | 5.00 | - | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAGG....................................................................................... | 22 | 1 | 15.00 | 15.00 | 6.00 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGT....................................................................................... | 21 | 1 | 14.00 | 18.00 | 8.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGATCTCTTGTTTTCTCTTCCAGT................................................. | 25 | 1 | 12.00 | 7.00 | 3.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 6.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTA......................................................................................... | 20 | 1 | 11.00 | 11.00 | 1.00 | 3.00 | - | 1.00 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGATCTCTTGTTTTCTCTTCC.................................................... | 22 | 1 | 9.00 | 9.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGGA...................................................................................... | 22 | 1 | 9.00 | 20.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCAGAAGAGAGACAGGATCTAGT....................................................................................... | 23 | 1 | 9.00 | 6.00 | 5.00 | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAGGA...................................................................................... | 23 | 1 | 8.00 | 15.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGATCTCTTGTTTTCTCTTC..................................................... | 21 | 1 | 7.00 | 7.00 | 2.00 | - | - | - | - | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGATCTCTTGTTTTCTCTTCCAG.................................................. | 24 | 1 | 7.00 | 7.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGTA...................................................................................... | 22 | 1 | 6.00 | 18.00 | - | 2.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCAGAAGAGAGACAGGATCTAG........................................................................................ | 22 | 1 | 6.00 | 6.00 | 3.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGGAT..................................................................................... | 23 | 1 | 5.00 | 20.00 | 1.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GAAGAGAGACAGGATCTAGT....................................................................................... | 20 | 4.00 | 0.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................TGGATCTCTTGTTTTCTCTTCCA................................................... | 23 | 1 | 4.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGGAA..................................................................................... | 23 | 1 | 4.00 | 20.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTA......................................................................................... | 19 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAGTT...................................................................................... | 23 | 1 | 4.00 | 49.00 | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAT........................................................................................ | 21 | 1 | 4.00 | 11.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCAGAAGAGAGACAGGATCTA......................................................................................... | 21 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAT........................................................................................ | 20 | 1 | 3.00 | 4.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGGT...................................................................................... | 22 | 1 | 3.00 | 20.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAGGT...................................................................................... | 23 | 1 | 3.00 | 15.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAGTA...................................................................................... | 23 | 1 | 3.00 | 49.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GAAGAGAGACAGGATCTA......................................................................................... | 18 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCT.......................................................................................... | 18 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCT.......................................................................................... | 19 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGATCTCTTGTTTTCTCTTCCAT.................................................. | 24 | 1 | 3.00 | 4.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGGTT..................................................................................... | 23 | 1 | 3.00 | 20.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................GCCAGAAGAGAGACAGGATCTAGT....................................................................................... | 24 | 3.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................TGGATCTCTTGTTTTCTCTT...................................................... | 20 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAGGG...................................................................................... | 23 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGATCTCTTGTTTTCTCTTCCAGTT................................................ | 26 | 1 | 2.00 | 7.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGGATCTCTTGTTTTCTCTTCCATT................................................. | 25 | 1 | 2.00 | 4.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAGGTT..................................................................................... | 24 | 1 | 2.00 | 15.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGAA...................................................................................... | 22 | 1 | 2.00 | 18.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAGA....................................................................................... | 22 | 1 | 2.00 | 49.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAGAA...................................................................................... | 23 | 1 | 1.00 | 49.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGGGAT.................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................AGACAGGATCTAGGGAAAGGA................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AGAGAGACAGGATCTACTTT..................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAGTAT..................................................................................... | 24 | 1 | 1.00 | 49.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCAGAAGAGAGACAGGATC........................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATTTA......................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGAAA..................................................................................... | 23 | 1 | 1.00 | 18.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCCAGT....................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................CCAGAAGAGAGACAGGAT............................................................................................ | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................GATCTCTTGTTTTCTCTTCCAGT................................................. | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGTT...................................................................................... | 22 | 1 | 1.00 | 18.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTATGA...................................................................................... | 22 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCAGAAGAGAGACAGGATCTAGG....................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTT......................................................................................... | 19 | 1 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................CACCACCACCACACAGTGGTT............................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................TGGATCTCTTGTTTTCTCTTCCAGA................................................. | 25 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................GGATCTCTTGTTTTCTCTTC..................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCAGAAGAGAGACAGGATCTAT........................................................................................ | 22 | 1 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AGAGAGACAGGATCTAGGGAAAGG................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGATAACGTGGAGGTCTAAACCTTC....................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAGGAC..................................................................................... | 24 | 1 | 1.00 | 15.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGGTA..................................................................................... | 23 | 1 | 1.00 | 20.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTATT....................................................................................... | 21 | 1 | 1.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTAGGTTT.................................................................................... | 25 | 1 | 1.00 | 15.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAGAGAGACAGGATCTAGGGTT.................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................GAAGAGAGACAGGATCTAGGGTA.................................................................................... | 23 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................GGAAAGGATGAATCTAAAA..................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................AGAGAGACAGGATCTAAA....................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................GCCAGAAGAGAGACAGGAT............................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GAAGAGAGACAGGATCTAGGAA..................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................CAGAAGAGAGACAGGATCTATTA...................................................................................... | 23 | 1 | 1.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCAGAAGAGAGACAGGATCT.......................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCAGAAGAGAGACAGGATCTAGGT...................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGAAGAGAGACAGGATC........................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGAAACGGATAGATTGTCAACCAGAGACCTGAACTCCTGAGCTAGGAAAGACTTGAGTATTCGCAGGCCACTTTTGATAACGTGGAGGTCTAAACCTTCGATTTTCTTTAAGCCCTACCACCACCACCACACAGTTACTGCCAGAAGAGAGACAGGATCTAGGGAAAGGATGAATCTGGATCTCTTGTTTTCTCTTCCAGGGTCACCGGGGCAACAGCCTGGACAGGAGGTCCCAGGGCGGCCCTCACCC .........................................................................................................................................(((...((((((((((((((((((((...........)))))))))..))))).))))))))).................................................. ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR059768(GSM562830) Treg_control. (spleen) | mjLiverWT1() Liver Data. (liver) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR059765(GSM562827) DN3_control. (thymus) | SRR525243(SRA056111/SRX170319) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR525244(SRA056111/SRX170320) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR525240(SRA056111/SRX170316) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR525241(SRA056111/SRX170317) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR525238(SRA056111/SRX170314) Second IgG control. (Blood) | SRR073954(GSM629280) total RNA. (blood) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR073955(GSM629281) total RNA. (blood) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR525242(SRA056111/SRX170318) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR525239(SRA056111/SRX170315) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR065056(SRR065056) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................CTTTAAGCCCTACCACC................................................................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |