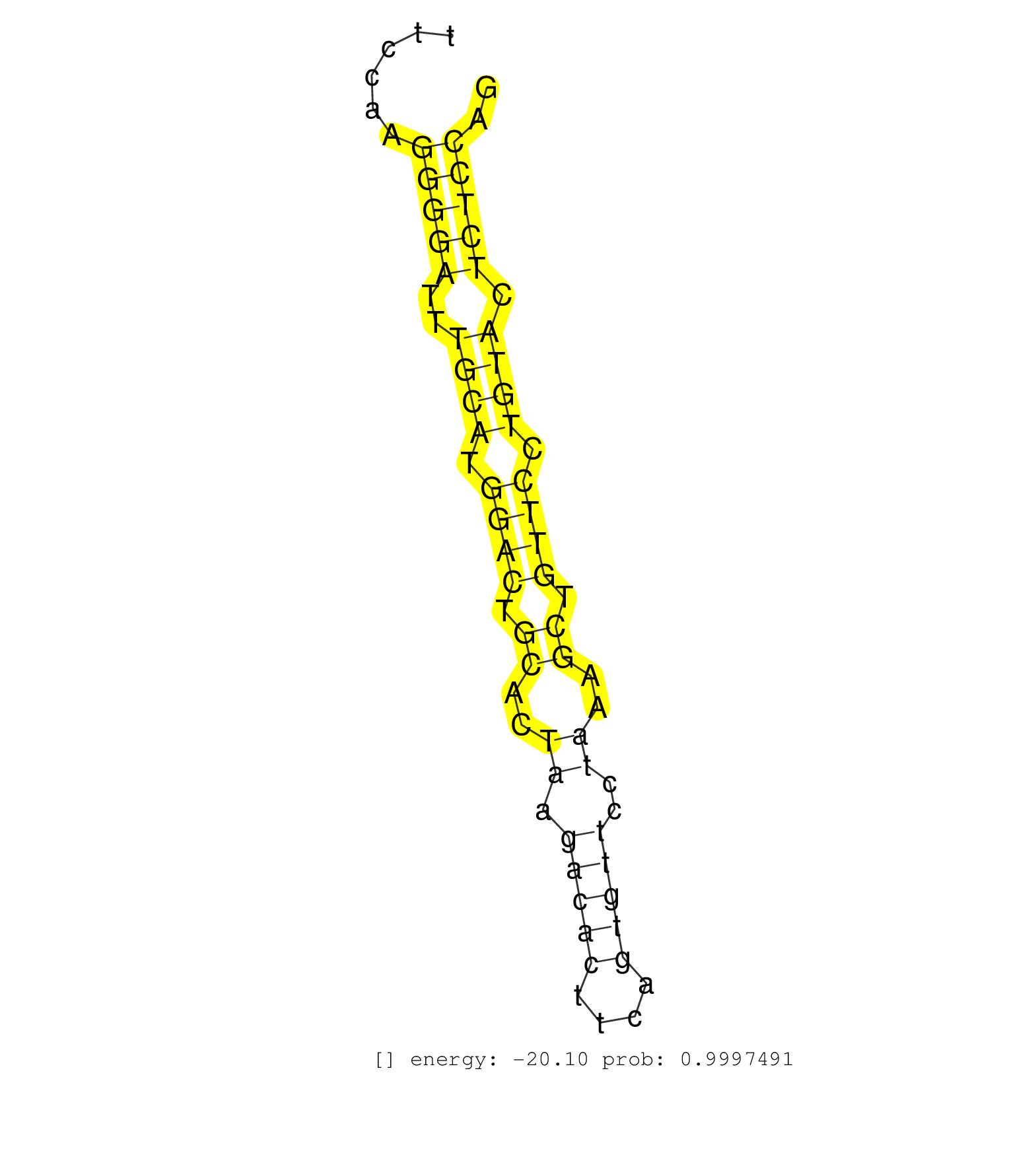
| (1) AGO.mut | (2) AGO1.ip | (17) AGO2.ip | (9) B-CELL | (46) BRAIN | (1) CELL-LINE | (1) DCR.mut | (2) DGCR8.mut | (8) EMBRYO | (4) ESC | (5) FIBROBLAST | (4) HEART | (2) KIDNEY | (9) LIVER | (1) LUNG | (2) LYMPH | (25) OTHER | (7) OTHER.mut | (1) OVARY | (1) PANCREAS | (2) SPLEEN | (17) TESTES | (1) TOTAL-RNA | (2) UTERUS |
| ATAGCTGTAGAGAATGTCAAGACTGCTCGGCATTATTGCTGTCTGCAGGCCACGTCACATGAGTGATTAATGTTCATTACCACAACAGGCCAGTCTGGCTGTGCTTATCCTGTAGTAAGCTAGGATTAGTTTCCAAGGGGATTTGCATGGACTGCACTAAGACACTTCAGTGTTCCTAAAGCTGTTCCTGTACTCTCCAGGTTCCGAGCCTCCCTCTATCCCTGCCCTGAGACTCCACAGGAAAGGAAAG ........................................................................................................................................(((((..((((.((((.((..((.(((((....)))))..))..)).)))).)))).))))).................................................... ..................................................................................................................................131..................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR553582(SRX182788) source: Brain. (Brain) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR206940(GSM723281) other. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | mjLiverWT2() Liver Data. (liver) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR073954(GSM629280) total RNA. (blood) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR206939(GSM723280) other. (brain) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | mjTestesWT3() Testes Data. (testes) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | mjTestesWT4() Testes Data. (testes) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037905(GSM510441) brain_rep2. (brain) | SRR069834(GSM304914) Analysis of small RNAs in murine neutrophils cultured in vitro by Solexa/Illumina genome analyzer. (blood) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR206941(GSM723282) other. (brain) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | SRR525238(SRA056111/SRX170314) Second IgG control. (Blood) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR553584(SRX182790) source: Heart. (Heart) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR206942(GSM723283) other. (brain) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR037906(GSM510442) brain_rep3. (brain) | SRR525237(SRA056111/SRX170313) Mus musculus domesticus miRNA sequencing. (Blood) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR065046(SRR065046) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | mjLiverWT1() Liver Data. (liver) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR065059(SRR065059) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037904(GSM510440) brain_rep1. (brain) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR346415(SRX098256) source: Testis. (Testes) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR042455(GSM539847) mouse plasma cells [09-002]. (blood) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | SRR095855BC6(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR065047(SRR065047) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR525239(SRA056111/SRX170315) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR553604(SRX182810) source: Spermatocytes. (Spermatocytes) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR037907(GSM510443) brain_rep4. (brain) | SRR553586(SRX182792) source: Testis. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR060845(GSM561991) total RNA. (brain) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR095855BC7(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR065056(SRR065056) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | GSM475281(GSM475281) total RNA. (testes) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR073955(GSM629281) total RNA. (blood) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR525241(SRA056111/SRX170317) Mus musculus domesticus miRNA sequencing. (ago1 Blood) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGT................................................. | 23 | 1 | 490.00 | 111.00 | 52.00 | 34.00 | 22.00 | 37.00 | 32.00 | 20.00 | - | 20.00 | 10.00 | - | 7.00 | 23.00 | - | 11.00 | 16.00 | 8.00 | 14.00 | - | 18.00 | 5.00 | - | 8.00 | 7.00 | - | 11.00 | 7.00 | - | 7.00 | 2.00 | 8.00 | 4.00 | 2.00 | 5.00 | 5.00 | 3.00 | 2.00 | - | 3.00 | 4.00 | 3.00 | 4.00 | 5.00 | 1.00 | 3.00 | 2.00 | 5.00 | 1.00 | 3.00 | 4.00 | 2.00 | 1.00 | 3.00 | 2.00 | 1.00 | - | - | 3.00 | - | 1.00 | - | 3.00 | 2.00 | - | - | 2.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | 2.00 | - | - | 2.00 | 2.00 | - | 2.00 | - | - | 1.00 | - | 1.00 | 2.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAG.................................................. | 22 | 1 | 111.00 | 111.00 | 11.00 | 12.00 | 1.00 | 4.00 | 3.00 | 3.00 | - | 8.00 | 6.00 | 5.00 | 9.00 | 2.00 | 6.00 | 4.00 | 1.00 | 1.00 | 1.00 | - | - | 2.00 | - | 2.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 3.00 | 1.00 | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCA................................................... | 21 | 1 | 105.00 | 105.00 | 3.00 | 2.00 | - | - | - | 11.00 | 17.00 | 3.00 | 4.00 | 10.00 | 4.00 | 3.00 | 6.00 | 4.00 | 1.00 | - | - | - | - | 3.00 | 7.00 | 2.00 | 3.00 | - | - | - | 7.00 | - | 2.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | 2.00 | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGA................................................. | 23 | 1 | 83.00 | 111.00 | 21.00 | 7.00 | 6.00 | 7.00 | 3.00 | - | - | 6.00 | - | - | - | - | - | 1.00 | - | 5.00 | - | - | - | 1.00 | - | 1.00 | - | 8.00 | - | - | - | - | - | - | - | - | 1.00 | 3.00 | - | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCC.................................................... | 20 | 1 | 72.00 | 72.00 | 1.00 | 2.00 | - | - | - | - | 14.00 | - | 3.00 | - | 7.00 | - | - | 2.00 | 6.00 | - | 1.00 | 12.00 | - | - | 5.00 | - | 1.00 | - | - | 1.00 | 4.00 | - | 4.00 | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCACT............................................................................................ | 23 | 1 | 35.00 | 35.00 | 1.00 | - | 11.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | 6.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCAC............................................................................................. | 22 | 1 | 19.00 | 19.00 | 3.00 | - | 1.00 | 1.00 | - | 2.00 | - | - | - | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGTA................................................ | 24 | 1 | 18.00 | 111.00 | 4.00 | - | 5.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCA.............................................................................................. | 21 | 1 | 13.00 | 13.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | 3.00 | - | - | 5.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCT...................................................... | 18 | 1 | 13.00 | 13.00 | - | - | - | - | - | - | 2.00 | - | - | 4.00 | - | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAT.................................................. | 22 | 1 | 9.00 | 105.00 | - | - | - | - | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGC................................................. | 23 | 1 | 8.00 | 111.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCACTAT.......................................................................................... | 25 | 1 | 8.00 | 7.00 | - | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGTT................................................ | 24 | 1 | 8.00 | 111.00 | - | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCACTA........................................................................................... | 24 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGC............................................................................................... | 20 | 1 | 6.00 | 6.00 | - | 1.00 | 2.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTC..................................................... | 19 | 1 | 5.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................AAGGGGATTTGCATGGACTGCAT............................................................................................. | 23 | 1 | 4.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCCGT................................................. | 23 | 1 | 4.00 | 72.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGAAT............................................... | 25 | 1 | 3.00 | 111.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................AGCTGTTCCTGTACTCTCCAGT................................................. | 22 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCACTT........................................................................................... | 24 | 1 | 2.00 | 35.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GGGATTTGCATGGACTGCACTAAG......................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAAA................................................. | 23 | 1 | 2.00 | 105.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCGCCA................................................... | 21 | 2.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGAA................................................ | 24 | 1 | 2.00 | 111.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................AAGGGGATTTGCATGGACTGCA.............................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TTTGCATGGACTGCACT............................................................................................ | 17 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCTA................................................... | 21 | 1 | 2.00 | 5.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCACTAA.......................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGTAA............................................... | 25 | 1 | 2.00 | 111.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCCG.................................................. | 22 | 1 | 1.00 | 72.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCCCCA................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCTA............................................................................................. | 22 | 1 | 1.00 | 6.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........GAGAATGTCAAGACTTATG.............................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................TTCCAAGGGGATTTGCATGGACTGT............................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCT.................................................... | 20 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGTTCCTGTACTCTCCAAC................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................GACACTTCAGTGTTCACGT....................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCACTAAGACA...................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TAGCTGTAGAGAATGTCAAGACTGC................................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGTAGA.............................................. | 26 | 1 | 1.00 | 111.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCCCC.................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCATT................................................. | 23 | 1 | 1.00 | 105.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAAAGA............................................... | 25 | 1 | 1.00 | 105.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGA............................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGAGT............................................... | 25 | 1 | 1.00 | 111.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................AAGGGGATTTGCATGGACTGCACTAAG......................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TTTGCATGGACTGCACA............................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCAT............................................................................................. | 22 | 1 | 1.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCATA............................................................................................ | 23 | 1 | 1.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................AAGCTAGGATTAGTTTA..................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCACTAG.......................................................................................... | 25 | 1 | 1.00 | 7.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCACTAAGAC....................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............GTCAAGACTGCTCGGCATT........................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........AGAGAATGTCAAGACTGCTCGGCATT........................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGGT................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGTC................................................ | 24 | 1 | 1.00 | 111.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCACTAAG......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........GAATGTCAAGACTGCTCGGCATT........................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................AAGGGGATTTGCATGGACTGTAT............................................................................................. | 23 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCTG.................................................. | 22 | 1 | 1.00 | 72.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......GTAGAGAATGTCAAGGTGC................................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................ATTAGTTTCCAAGGGACA............................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ...............GTCAAGACTGCTCGGGGT......................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................CTAAAGCTGTTCCTGTTTGT....................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................GCTGTTCCTGTACTCTCCAGT................................................. | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCAA............................................................................................. | 22 | 1 | 1.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCATT............................................................................................ | 23 | 1 | 1.00 | 13.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAAAA................................................ | 24 | 1 | 1.00 | 105.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GGGGATTTGCATGGACTGCACTA........................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGGGGATTTGCATGGACTGCCC............................................................................................. | 22 | 1 | 1.00 | 6.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGTTT............................................... | 25 | 1 | 1.00 | 111.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CTGTTCCTGTACTCTCCAG.................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TAATGTTCATTACCAAGGC.................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCAAGT................................................. | 23 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................AAAGCTGTTCCTGTACTCTCCAGGT................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGTTCCTGTACTCTCCAGT................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................AAGCTGTTCCTGTACTCTCCAGG................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATAGCTGTAGAGAATGTCAAGACTGCTCGGCATTATTGCTGTCTGCAGGCCACGTCACATGAGTGATTAATGTTCATTACCACAACAGGCCAGTCTGGCTGTGCTTATCCTGTAGTAAGCTAGGATTAGTTTCCAAGGGGATTTGCATGGACTGCACTAAGACACTTCAGTGTTCCTAAAGCTGTTCCTGTACTCTCCAGGTTCCGAGCCTCCCTCTATCCCTGCCCTGAGACTCCACAGGAAAGGAAAG ........................................................................................................................................(((((..((((.((((.((..((.(((((....)))))..))..)).)))).)))).))))).................................................... ..................................................................................................................................131..................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR553582(SRX182788) source: Brain. (Brain) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR206940(GSM723281) other. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | mjLiverWT2() Liver Data. (liver) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR073954(GSM629280) total RNA. (blood) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR206939(GSM723280) other. (brain) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | mjTestesWT3() Testes Data. (testes) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | mjTestesWT4() Testes Data. (testes) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037905(GSM510441) brain_rep2. (brain) | SRR069834(GSM304914) Analysis of small RNAs in murine neutrophils cultured in vitro by Solexa/Illumina genome analyzer. (blood) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR206941(GSM723282) other. (brain) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | SRR525238(SRA056111/SRX170314) Second IgG control. (Blood) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR553584(SRX182790) source: Heart. (Heart) | SRR390297(GSM849855) cell line: NIH-3T3cell type: fibroblastip ant. (fibroblast) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR206942(GSM723283) other. (brain) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR037906(GSM510442) brain_rep3. (brain) | SRR525237(SRA056111/SRX170313) Mus musculus domesticus miRNA sequencing. (Blood) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR065046(SRR065046) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | mjLiverWT1() Liver Data. (liver) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR065059(SRR065059) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037904(GSM510440) brain_rep1. (brain) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR346415(SRX098256) source: Testis. (Testes) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR042455(GSM539847) mouse plasma cells [09-002]. (blood) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | SRR095855BC6(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR065047(SRR065047) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR525239(SRA056111/SRX170315) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR553604(SRX182810) source: Spermatocytes. (Spermatocytes) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR037907(GSM510443) brain_rep4. (brain) | SRR553586(SRX182792) source: Testis. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR060845(GSM561991) total RNA. (brain) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR095855BC7(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR065056(SRR065056) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | GSM475281(GSM475281) total RNA. (testes) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR073955(GSM629281) total RNA. (blood) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR525241(SRA056111/SRX170317) Mus musculus domesticus miRNA sequencing. (ago1 Blood) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................TAAGCTAGGATTAGTGCCC.................................................................................................................... | 19 | 16.75 | 0.00 | - | - | - | - | - | 2.25 | 1.50 | - | 0.50 | - | - | - | - | - | - | - | - | 6.00 | - | 0.25 | 1.50 | - | - | - | - | - | 0.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.75 | - | - | - | - | - | - | - | 0.25 | 1.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.75 | 0.25 | - | |
| ...................................................................................................................TAAGCTAGGATTAGTGC...................................................................................................................... | 17 | 4.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.75 | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | |
| ...................................................................................................................TAAGCTAGGATTAGT........................................................................................................................ | 15 | 4 | 2.50 | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................CTCCCTCTATCCCTGCCGAG..................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................GTAAGCTAGGATTAGTAC...................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................CTGCCCTGAGACTCCACCT.......... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................ATTGCTGTCTGCAGGCCACTT................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................TCCGAGCCTCCCTCTCTC.............................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................GTTCCTAAAGCTGTTCCTGT........................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................CTCTCCAGGTTCCGAGCCT....................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................GTAAGCTAGGATTAGT........................................................................................................................ | 16 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................GTAAGCTAGGATTAGTA....................................................................................................................... | 17 | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................TAAGCTAGGATTAGTG....................................................................................................................... | 16 | 0.50 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................TAAGCTAGGATTAGTGCC..................................................................................................................... | 18 | 0.25 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................TAAGCTAGGATTAGTGA...................................................................................................................... | 17 | 0.25 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................TAAGCTAGGATTAGTGCCA.................................................................................................................... | 19 | 0.25 | 0.00 | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................TAAGCTAGGATTAGTGTGC.................................................................................................................... | 19 | 0.25 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................TAAGCTAGGATTAGTGCCT.................................................................................................................... | 19 | 0.25 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |