| (1) AGO1.ip | (11) AGO2.ip | (1) B-CELL | (34) BRAIN | (5) CELL-LINE | (1) DCR.mut | (2) DGCR8.mut | (10) EMBRYO | (2) ESC | (3) FIBROBLAST | (1) HEART | (1) KIDNEY | (2) LIVER | (4) LYMPH | (11) OTHER | (7) OTHER.mut | (2) PIWI.ip | (2) SKIN | (4) SPLEEN | (16) TESTES | (2) THYMUS | (4) TOTAL-RNA | (2) UTERUS |
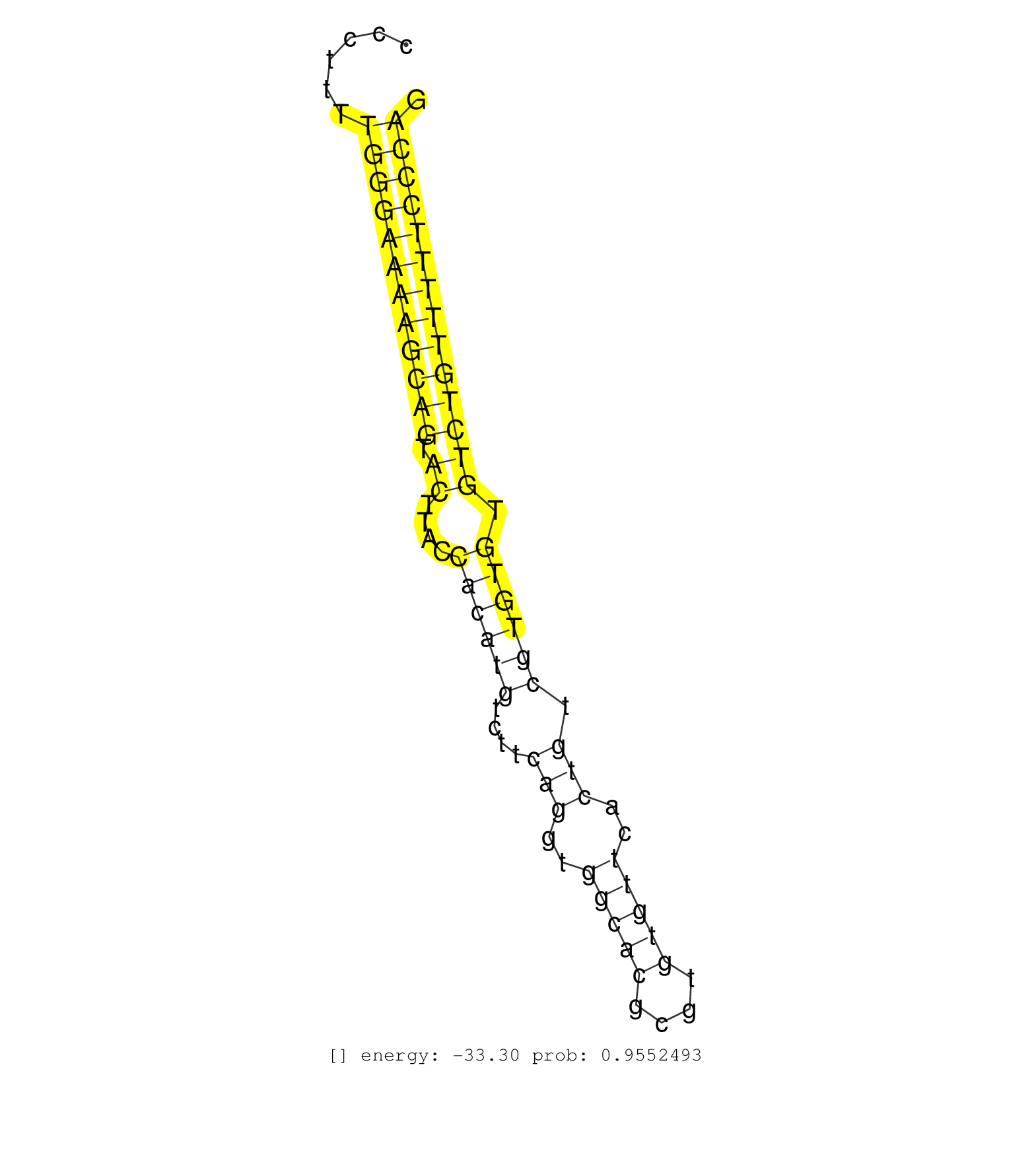
| GTGAACTCAGCGAATCAATTGCCTGTGGGTGGTTACTGTCGACCCAGCAGCAAAAAGACAAAGGAAGGTGAGAGAGTAGAGGCAGAATAGTAGAGACAGGGGAGAAGACCCAAGGTGTCCCTTTTGGGAAAAGCAGTACTTACCACATGTCTTCAGGTGGCACGCGTGTGTTCACTGTCGTGTGTGTCTGTTTTTCCCAGGTATCATTGACAGCACTGCTGGGGAACAGCGGCAGGTAGTGGCTGTCACC ............................................................................................................................((((((((((((.((....((((((....(((..(((((....)))))..))).)))))).))))))))))))))................................................... ......................................................................................................................119..............................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR073955(GSM629281) total RNA. (blood) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | GSM510444(GSM510444) brain_rep5. (brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR073954(GSM629280) total RNA. (blood) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR206942(GSM723283) other. (brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR037902(GSM510438) testes_rep3. (testes) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR206941(GSM723282) other. (brain) | SRR037905(GSM510441) brain_rep2. (brain) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR065049(SRR065049) Tissue-specific Regulation of Mouse MicroRNA . (liver) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR037922(GSM510460) e12p5_rep4. (embryo) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR037903(GSM510439) testes_rep4. (testes) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR059768(GSM562830) Treg_control. (spleen) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR553584(SRX182790) source: Heart. (Heart) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR346415(SRX098256) source: Testis. (Testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR059765(GSM562827) DN3_control. (thymus) | SRR059771(GSM562833) CD4_control. (spleen) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR037906(GSM510442) brain_rep3. (brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR206939(GSM723280) other. (brain) | GSM416732(GSM416732) MEF. (cell line) | SRR553604(SRX182810) source: Spermatocytes. (Spermatocytes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACC.......................................................................................................... | 21 | 1 | 43.00 | 43.00 | - | 8.00 | 3.00 | 2.00 | 4.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | 1.00 | 1.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 2.00 | 1.00 | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACCA......................................................................................................... | 22 | 1 | 31.00 | 31.00 | - | - | 2.00 | - | 1.00 | - | - | 4.00 | - | - | 2.00 | 1.00 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | 3.00 | - | 1.00 | - | - | - | 1.00 | - | 2.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGC................................................. | 21 | 21.00 | 0.00 | - | 11.00 | - | 2.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACCACATGTCTTCAGGTGGCACGCGTGTGTTCACTGTCGTGTGTGTCTGTTTTTCCCAGT................................................. | 78 | 19.00 | 0.00 | 19.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTAC........................................................................................................... | 20 | 1 | 19.00 | 19.00 | - | 1.00 | 3.00 | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................TGGGAAAAGCAGTACTTACCACATGTCTTCAGGTGGCACGCGTGTGTTCACTGTCGTGTGTGTCTGTTTTTCCCAGT................................................. | 77 | 15.00 | 0.00 | 15.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACCACAT...................................................................................................... | 25 | 1 | 13.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGTT................................................ | 22 | 11.00 | 0.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGT................................................. | 21 | 11.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTA............................................................................................................ | 19 | 1 | 10.00 | 10.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACCACA....................................................................................................... | 24 | 1 | 10.00 | 10.00 | - | - | - | 1.00 | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACCAC........................................................................................................ | 23 | 1 | 9.00 | 9.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACCACATG..................................................................................................... | 26 | 1 | 7.00 | 7.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACA.......................................................................................................... | 21 | 1 | 4.00 | 19.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGTTT............................................... | 23 | 3.00 | 0.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGTTG............................................... | 23 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................TGGGAAAAGCAGTACTTACC.......................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACCACATGTCT.................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGTA................................................ | 22 | 2.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................TTTTGGGAAAAGCAGTACTTACC.......................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGGGAAAAGCAGTACTTACCACAT...................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGAAC............................................... | 23 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACCACATGT.................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CGTGTGTGTCTGTTTTTCCCAGT................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................TGTGTCTGTTTTTCCCAGTTT............................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CGTGTGTGTCTGTTTTTCCCAGA................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACCAT........................................................................................................ | 23 | 1 | 1.00 | 31.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACAC......................................................................................................... | 22 | 1 | 1.00 | 19.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TTGGGAAAAGCAGTACATA............................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................TTTGGGAAAAGCAGTACTTACC.......................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................CAGCACTGCTGGGGAACAGCGGCAGGT............. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTATCA......................................................................................................... | 22 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACCACATGTCTTCAGGTGGCACGCGTGTGTTCACTGTCGTGTGTGTCTGTTTTTCCCATT................................................. | 78 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................TGTGTCTGTTTTTCCCAGAAC............................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................GTGTGTGTCTGTTTTTCCCAGA................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................TGGGAAAAGCAGTACTTACCACA....................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGCTGGGGAACAGCGGCAGG.............. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGCCA............................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGCAG............................................... | 23 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACCACGT...................................................................................................... | 25 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCAGTT................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................TGTGTCTGTTTTTCCCAGAGA............................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACCACATGTCTTCAGGTGGCACGCGTGTGTTCACTGTCGTGTGTGTCTGTTTTTCCCAGA................................................. | 78 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGAAA............................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................CTGCTGGGGAACAGCGGCAGG.............. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGTCTTCAGGTGGCAGTGA.................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................TTTTGGGAAAAGCAGTACTTAC........................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GCTGGGGAACAGCGGCAGGT............. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGGGAAAAGCAGTACTTACCA......................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGAT................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................TTGGGAAAAGCAGTACTTACCACATGTCTTCAGGTGGCACGCGTGTGTTCACTGTCGTGTGTGTCTGTTTTTCCTAGT................................................. | 78 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................TTGGGAAAAGCAGTACTT............................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................ATTGACAGCACTGCTG............................. | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AGGTGAGAGAGTAGAAC........................................................................................................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGTTA............................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGAGA............................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TGTGTGTCTGTTTTTCCCAGGC................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CGTGTGTGTCTGTTTTTCCCA................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................................................................AGCGGCAGGTAGTGGCT...... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................CAGGTAGTGGCTGTCAC. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ....................................................................TGAGAGAGTAGAGGCA...................................................................................................................................................................... | 16 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| GTGAACTCAGCGAATCAATTGCCTGTGGGTGGTTACTGTCGACCCAGCAGCAAAAAGACAAAGGAAGGTGAGAGAGTAGAGGCAGAATAGTAGAGACAGGGGAGAAGACCCAAGGTGTCCCTTTTGGGAAAAGCAGTACTTACCACATGTCTTCAGGTGGCACGCGTGTGTTCACTGTCGTGTGTGTCTGTTTTTCCCAGGTATCATTGACAGCACTGCTGGGGAACAGCGGCAGGTAGTGGCTGTCACC ............................................................................................................................((((((((((((.((....((((((....(((..(((((....)))))..))).)))))).))))))))))))))................................................... ......................................................................................................................119..............................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR073955(GSM629281) total RNA. (blood) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | GSM510444(GSM510444) brain_rep5. (brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR073954(GSM629280) total RNA. (blood) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR206942(GSM723283) other. (brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR037902(GSM510438) testes_rep3. (testes) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR685339(GSM1079783) "Small RNAs (15-50 nts in length) from immort. (cell line) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR206941(GSM723282) other. (brain) | SRR037905(GSM510441) brain_rep2. (brain) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR065049(SRR065049) Tissue-specific Regulation of Mouse MicroRNA . (liver) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR037922(GSM510460) e12p5_rep4. (embryo) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR037903(GSM510439) testes_rep4. (testes) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR059768(GSM562830) Treg_control. (spleen) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR553584(SRX182790) source: Heart. (Heart) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR346415(SRX098256) source: Testis. (Testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR059765(GSM562827) DN3_control. (thymus) | SRR059771(GSM562833) CD4_control. (spleen) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR037906(GSM510442) brain_rep3. (brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR206939(GSM723280) other. (brain) | GSM416732(GSM416732) MEF. (cell line) | SRR553604(SRX182810) source: Spermatocytes. (Spermatocytes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................................GGTAGTGGCTGTCACCCC | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................................GCTGGGGAACAGCGGCA................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................GTGTGTTCACTGTCGTT.................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................TTTTTCCCAGGTATC............................................. | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - |