| (1) B-CELL | (2) BRAIN | (10) BREAST | (15) CELL-LINE | (3) CERVIX | (1) FIBROBLAST | (1) HEART | (3) LIVER | (3) OTHER | (2) SKIN |
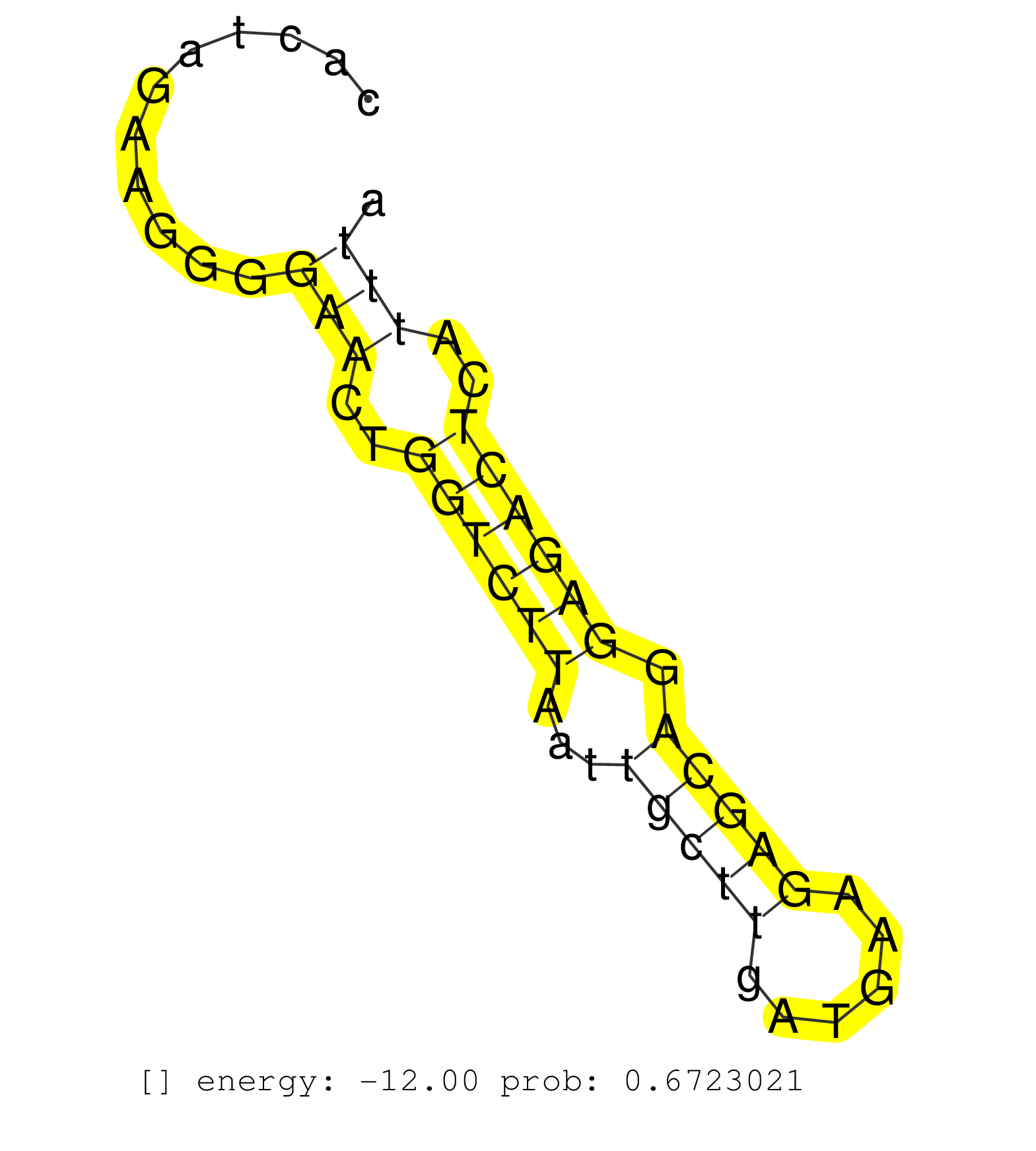
| TGTCCCCGTTGGATAATGAGGACAAGGGCTGGGGGCTCCGGTGGTTTGCGGCAGGGACTTGATCACATCCTTCTGCTGTGGCCCCATTGCCTCTGGCTGGAGTTGACCCTTCTGACAAGTGTCCTCAGAAAGACAGGGATCACCGGCACCTCCCAATATCAACCCCAGGCAGCACAGACACAAACCCCACATCCAGAGCCAACTCCAGGAGCAGAGACACCCCAACACTCTGGGGGACCCCAACCGTGATAACTCCCCACTGGAATCCGCCCCAGAG .....................................................................................((((......(((((...((((((((((((.........))))).....)))).))))))))......))))........................................................................................................................ .....................................................................................86.........................................................................161.................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | TAX577741(Rovira) total RNA. (breast) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR189782 | TAX577740(Rovira) total RNA. (breast) | SRR191629(GSM715739) 5genomic small RNA (size selected RNA from to. (breast) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR189784 | SRR037935(GSM510473) 293cand3. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR191544(GSM715654) 126genomic small RNA (size selected RNA from . (breast) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR040039(GSM532924) G531T. (cervix) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR191548(GSM715658) 101genomic small RNA (size selected RNA from . (breast) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR343335 | SRR040032(GSM532917) G603N. (cervix) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | TAX577738(Rovira) total RNA. (breast) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | TAX577745(Rovira) total RNA. (breast) | SRR191497(GSM715607) 17genomic small RNA (size selected RNA from t. (breast) | SRR040015(GSM532900) G623T. (cervix) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................ATCACCGGCACCTCCACCA........................................................................................................................ | 19 | 36.00 | 0.00 | 4.00 | 3.00 | 2.00 | 3.00 | 2.00 | 2.00 | - | 1.00 | 1.00 | 2.00 | 1.00 | - | 2.00 | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | |
| ...........................................................................................................................................TCACCGGCACCTCCCCCA........................................................................................................................ | 18 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................ATCACCGGCACCTCCACC......................................................................................................................... | 18 | 3.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .........................................................................................................................................GATCACCGGCACCTCCACCA........................................................................................................................ | 20 | 3.00 | 0.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................CGGCAGGGACTTGAT...................................................................................................................................................................................................................... | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................CACTCTGGGGGACCCCA................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AGGGCTGGGGGCTCCGGGAC......................................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................ATCACCGGCACCTCCACAA........................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................CTCTGGCTGGAGTTGACCC........................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GATCACCGGCACCTCCACC......................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................ATCACCGGCACCTCCACT......................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................................CACCCCAACACTCTGGCACT........................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................ATCACCGGCACCTCCACTA........................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................TCCTCAGAAAGACAGTGG.......................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ............................CTGGGGGCTCCGGTG.......................................................................................................................................................................................................................................... | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |
| TGTCCCCGTTGGATAATGAGGACAAGGGCTGGGGGCTCCGGTGGTTTGCGGCAGGGACTTGATCACATCCTTCTGCTGTGGCCCCATTGCCTCTGGCTGGAGTTGACCCTTCTGACAAGTGTCCTCAGAAAGACAGGGATCACCGGCACCTCCCAATATCAACCCCAGGCAGCACAGACACAAACCCCACATCCAGAGCCAACTCCAGGAGCAGAGACACCCCAACACTCTGGGGGACCCCAACCGTGATAACTCCCCACTGGAATCCGCCCCAGAG .....................................................................................((((......(((((...((((((((((((.........))))).....)))).))))))))......))))........................................................................................................................ .....................................................................................86.........................................................................161.................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | TAX577741(Rovira) total RNA. (breast) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR189782 | TAX577740(Rovira) total RNA. (breast) | SRR191629(GSM715739) 5genomic small RNA (size selected RNA from to. (breast) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR189784 | SRR037935(GSM510473) 293cand3. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR191544(GSM715654) 126genomic small RNA (size selected RNA from . (breast) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR040039(GSM532924) G531T. (cervix) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR191548(GSM715658) 101genomic small RNA (size selected RNA from . (breast) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR343335 | SRR040032(GSM532917) G603N. (cervix) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | TAX577738(Rovira) total RNA. (breast) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | TAX577745(Rovira) total RNA. (breast) | SRR191497(GSM715607) 17genomic small RNA (size selected RNA from t. (breast) | SRR040015(GSM532900) G623T. (cervix) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................CATCCTTCTGCTGTGGCCA................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .GTCCCCGTTGGATAATCCA................................................................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................GGGATCACCGGCACCTCG............................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................AGGACACTTGTCAGAAGG........................................................................................................................................................ | 18 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |
| .............................................................CAGCAGAAGGATGTGAT....................................................................................................................................................................................................... | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - |