| (12) B-CELL | (2) BREAST | (3) CELL-LINE | (2) CERVIX | (1) FIBROBLAST | (2) HEART | (1) LIVER | (1) OTHER | (4) SKIN |
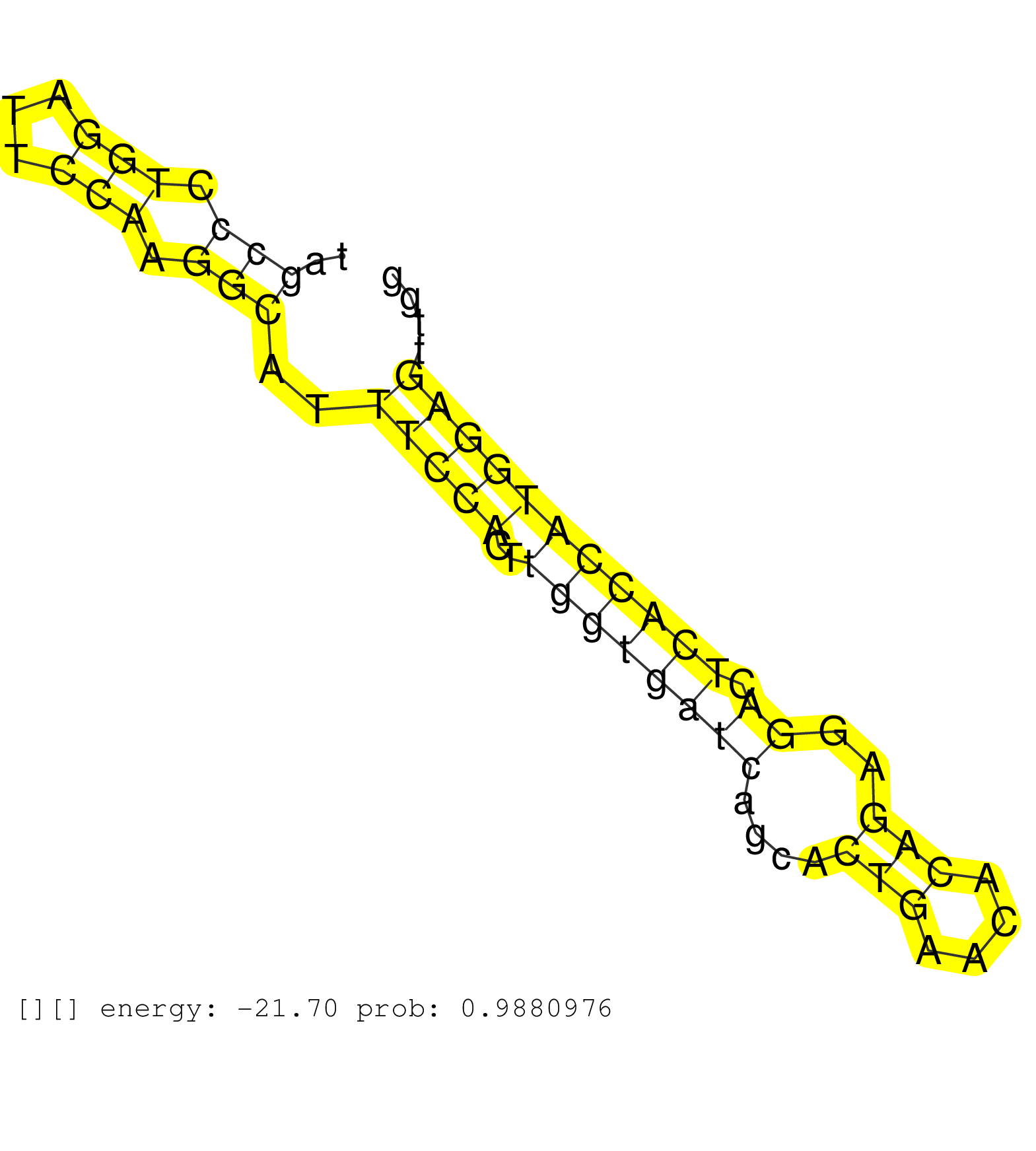
| AATGACCCTGCCTTGGTCACATTGATCAGTGTCCTGAGGAAGCCCAGTGGAACCTGGGGAGTGTTTTCCAGTCAGGCTCAGGGCAGAGACCTCCGTGGAAATCTCTGATTAGAATAGGTTTGGGATTCAGACTTGGGCCAAGAGGGAGTCTCACCCATGGGAGGGTCCTTAAATCCTGACAGTTTTTACAGCTGTTCTCCCTCCTCTGGCAAAACTGTGCACATCTGACTCAGAACTGATCCAGCTGGCCCTTACTTGCTAATCAATTTTTCTTCTCTCTACACTTCATTC ...............................................................................................................((.(((.((((((..(((((..((.((((.(((((((....((((......))))...........(((((((.....))))))))))))))...)))).........))..))))))))))).))).)).................................................. ..............................................................................................................111...............................................................................................................................241................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | TAX577746(Rovira) total RNA. (breast) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR040036(GSM532921) G243N. (cervix) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577589(Rovira) total RNA. (breast) | GSM532879(GSM532879) G659N. (cervix) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR189784 | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................ACATCTGACTCAGAACTGGCA.................................................. | 21 | 67.00 | 0.00 | 57.00 | 5.00 | 2.00 | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................ATCTGACTCAGAACTGGCA.................................................. | 19 | 48.00 | 0.00 | 42.00 | 3.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................TCTGACTCAGAACTGGCA.................................................. | 18 | 43.00 | 0.00 | 38.00 | 2.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................TCTGACTCAGAACTGGCAC................................................. | 19 | 23.00 | 0.00 | 12.00 | 4.00 | 1.00 | 2.00 | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................................ACATCTGACTCAGAACTGGCAC................................................. | 22 | 22.00 | 0.00 | 19.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................ATCTGACTCAGAACTGGCAC................................................. | 20 | 20.00 | 0.00 | 12.00 | - | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................CATCTGACTCAGAACTGGCA.................................................. | 20 | 18.00 | 0.00 | 12.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ............................................................................................................................................................................................................................ACATCTGACTCAGAACTGGC................................................... | 20 | 10.00 | 0.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................CATCTGACTCAGAACTGGCAC................................................. | 21 | 7.00 | 0.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................ATCTGACTCAGAACTGGC................................................... | 18 | 5.00 | 0.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................AAGCCCAGTGGAACCACCA......................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................TCTGACTCAGAACTGGCAG................................................. | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................CATCTGACTCAGAACTGGC................................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................AGGTTTGGGATTCAGGG............................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ............................................................................CTCAGGGCAGAGACCTCTGA................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................ACATTGATCAGTGTCAAT............................................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................ATCTGACTCAGAACTGACAC................................................. | 20 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................................ACATCTGACTCAGAACTGCCA.................................................. | 21 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................................ACATCTGACTCAGAACTGGCC.................................................. | 21 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................................ACATCTGACTCAGAACTGGCAA................................................. | 22 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................TGACAGTTTTTACAGACAT................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| .....................................................................................................................................................CTCACCCATGGGAGGG.............................................................................................................................. | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| AATGACCCTGCCTTGGTCACATTGATCAGTGTCCTGAGGAAGCCCAGTGGAACCTGGGGAGTGTTTTCCAGTCAGGCTCAGGGCAGAGACCTCCGTGGAAATCTCTGATTAGAATAGGTTTGGGATTCAGACTTGGGCCAAGAGGGAGTCTCACCCATGGGAGGGTCCTTAAATCCTGACAGTTTTTACAGCTGTTCTCCCTCCTCTGGCAAAACTGTGCACATCTGACTCAGAACTGATCCAGCTGGCCCTTACTTGCTAATCAATTTTTCTTCTCTCTACACTTCATTC ...............................................................................................................((.(((.((((((..(((((..((.((((.(((((((....((((......))))...........(((((((.....))))))))))))))...)))).........))..))))))))))).))).)).................................................. ..............................................................................................................111...............................................................................................................................241................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR015363(GSM380328) Germinal Center B cell (GC40). (B cell) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | TAX577746(Rovira) total RNA. (breast) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR040036(GSM532921) G243N. (cervix) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577589(Rovira) total RNA. (breast) | GSM532879(GSM532879) G659N. (cervix) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR189784 | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................GTTCTCCCTCCTCTGTCTG............................................................................... | 19 | 6.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................CTGGCAAAACTGTGCGTA.................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................TTGGGCCAAGAGGGACGG............................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................................................AACTGATCCAGCTGGCCGT....................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................................................CAGAACTGATCCAGCTCAC.......................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................CCACGGAGGTCTCTGCC................................................................................................................................................................................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ...........................................................................................................................................................................................AGAGGAGGGAGAACAGCTGT.................................................................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| .........................................................................................................................................................................................................................TCTGAGTCAGATGTGCA......................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |