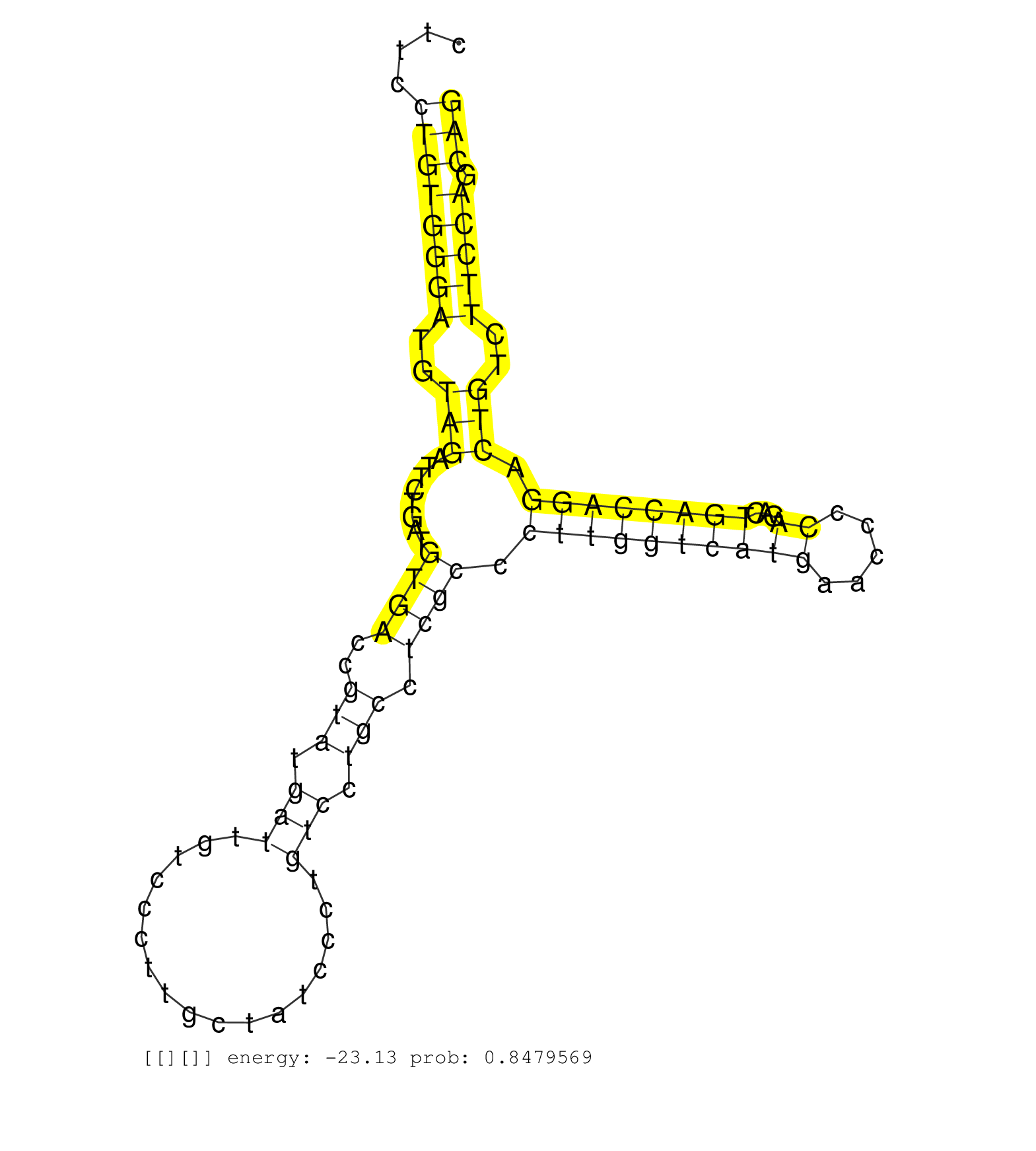
| (2) AGO2.ip | (6) B-CELL | (3) BRAIN | (10) BREAST | (36) CELL-LINE | (7) HEART | (2) HELA | (1) KIDNEY | (7) LIVER | (2) OTHER | (2) RRP40.ip | (30) SKIN | (1) TESTES | (2) UTERUS | (2) XRN.ip |
| CTGACTGATCTTGTGTGCCTGAGATGTAGGACTTCCTCTGAGAGTGATCATGGAGAAACGGTTTGGGTGGCTGGTGGGGAAGGTACTGGCTTCCTGTGGGATGTAGATTCTGATGTGACCGTATGATTGTCCCTTGCTATCCCTGTCCTGCCTCGCCCTTGGTCATGAACCCCAGACTGACCAGGACTGTCTTCCAGCAGGAGGCCACTGTCTGCAGCTCCCGTGAAGATGTCCACTCCAGACCCACCCC .............................................................................................((((((((..(((........((((..(((.(((.................))).))).)))).((((((((((.....))...)))))))).)))..))))).))).................................................. .........................................................................................90............................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR553575(SRX182781) source: Kidney. (Kidney) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR038858(GSM458541) MEL202. (cell line) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553576(SRX182782) source: Testis. (testes) | SRR038857(GSM458540) D20. (cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | TAX577738(Rovira) total RNA. (breast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR038862(GSM458545) MM472. (cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | TAX577580(Rovira) total RNA. (breast) | SRR207117(GSM721079) Whole cell RNA. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191397(GSM715507) 30genomic small RNA (size selected RNA from t. (breast) | TAX577588(Rovira) total RNA. (breast) | SRR037935(GSM510473) 293cand3. (cell line) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR207111(GSM721073) Whole cell RNA. (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577741(Rovira) total RNA. (breast) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | TAX577590(Rovira) total RNA. (breast) | SRR038861(GSM458544) MM466. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | TAX577589(Rovira) total RNA. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR038853(GSM458536) MELB. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038856(GSM458539) D11. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | TAX577745(Rovira) total RNA. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR553574(SRX182780) source: Heart. (Heart) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAGCAG.................................................. | 28 | 1 | 109.00 | 109.00 | - | 16.00 | 17.00 | 5.00 | - | - | 2.00 | 5.00 | 3.00 | 1.00 | 2.00 | 2.00 | 1.00 | - | 3.00 | 1.00 | 3.00 | 2.00 | 2.00 | 2.00 | - | - | 3.00 | 1.00 | 1.00 | 3.00 | - | 2.00 | - | - | 1.00 | 1.00 | - | 1.00 | 2.00 | 1.00 | - | 2.00 | - | - | 1.00 | - | 1.00 | 1.00 | 2.00 | - | 2.00 | 2.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAGAA................................................... | 27 | 1 | 21.00 | 9.00 | - | - | - | - | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAGCA................................................... | 27 | 1 | 11.00 | 11.00 | - | - | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAG..................................................... | 25 | 1 | 9.00 | 9.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAGA.................................................... | 26 | 1 | 9.00 | 9.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAGC.................................................... | 26 | 1 | 7.00 | 7.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAGCAGAA................................................ | 30 | 1 | 6.00 | 109.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................CCAGACTGACCAGGACTGTCTTCCAGAAA.................................................. | 29 | 1 | 6.00 | 1.00 | - | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGAGGCCACTGTCTGCAGCT............................... | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................CCAGACTGACCAGGACTGTCTTCCAGCAG.................................................. | 29 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................AGACTGACCAGGACTGTCTTCCAGA.................................................... | 25 | 3.00 | 0.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAGAAA.................................................. | 28 | 1 | 3.00 | 9.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................AAGATGTCCACTCCAGA........ | 17 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAGCAT.................................................. | 28 | 1 | 2.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTGGGATGTAGATTCTGATG....................................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................CTGAGAGTGATCATGGA.................................................................................................................................................................................................... | 17 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................AGACTGACCAGGACTGTCTTCCAGCAG.................................................. | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAGAT................................................... | 27 | 1 | 2.00 | 9.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GTAGATTCTGATGTGACC.................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TGGCTGGTGGGGAAGCGC..................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................CTTGGTCATGAACCCCAGACTGACC.................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................CCCAGACTGACCAGGACTGTCTTCCAGCAG.................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................AGGCCACTGTCTGCAGCTCCCGTGAAGATGTC................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CACTGTCTGCAGCTCCCGTGAAGATGTC................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTT......................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CTGTGGGATGTAGATTCTGATGTGA.................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TCTGATGTGACCGTATGATTGT........................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................CCCAGACTGACCAGGACTGTCTTCCAGCAGGAG............................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................AGAGTGATCATGGAGAAACGGTT........................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCACCAG.................................................. | 28 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................AGATGTAGGACTTCCTCTGAGAGTGATCATG...................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CACTGTCTGCAGCTCCCGTGAAGA..................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGACCAGGACTGTCTTCCAGAAA.................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTC........................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGAGGCCACTGTCTGCAGCTC.............................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................CCCCAGACTGACCAGGACTGTCTTCCAGA.................................................... | 29 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................GAGATGTAGGACTTCCTCTGAGAGT............................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAGTT................................................... | 27 | 1 | 1.00 | 9.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCC....................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................GTCTGCAGCTCCCGTGAAGATG................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGTGGGATGTAGATTCTGATGTGA.................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAGAAAA................................................. | 29 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GAGGCCACTGTCTGCAGCTCCCGTGAAGATG................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TTTGGGTGGCTGGTGGG............................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................GACCAGGACTGTCTTCCAGCAGGAGGC............................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GAGGCCACTGTCTGCAGCTCCCGTGAAGATGTC................. | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CTGACCAGGACTGTCTTCC....................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................AGACTGACCAGGACTGTCTTCCA...................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GCTTCCTGTGGGATGTAGATTCTGATGTGA.................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................GGGTGGCTGGTGGGGGTCC....................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................TGGGATGTAGATTCTGATG....................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAGAG................................................... | 27 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GAGGCCACTGTCTGCAGCT............................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCATA.................................................... | 26 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCAGCAGA................................................. | 29 | 1 | 1.00 | 109.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................AGGACTGTCTTCCAGCAGAAA............................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................GGATGTAGATTCTGATG....................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................TCATGGAGAAACGGTTTGGGTGGCTGGTGG............................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CAGGACTGTCTTCCAGCAG.................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................CAGCAGGAGGCCACTGTCTGCAGCT............................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................CCCAGACTGACCAGGACTGTCTTCCAG..................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................ACTTCCTCTGAGAGTGAGAG........................................................................................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................ACTGACCAGGACTGTCTTCCAG..................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGACTGACCAGGACTGTCTTCCACCCG.................................................. | 28 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............GTGTGCCTGAGATGTGATG........................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................CCAGACTGACCAGGACTGTCTTCCAG..................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................AAGGTACTGGCTTCC............................................................................................................................................................ | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - |
| ................GCCTGAGATGTAGGA........................................................................................................................................................................................................................... | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |
| CTGACTGATCTTGTGTGCCTGAGATGTAGGACTTCCTCTGAGAGTGATCATGGAGAAACGGTTTGGGTGGCTGGTGGGGAAGGTACTGGCTTCCTGTGGGATGTAGATTCTGATGTGACCGTATGATTGTCCCTTGCTATCCCTGTCCTGCCTCGCCCTTGGTCATGAACCCCAGACTGACCAGGACTGTCTTCCAGCAGGAGGCCACTGTCTGCAGCTCCCGTGAAGATGTCCACTCCAGACCCACCCC .............................................................................................((((((((..(((........((((..(((.(((.................))).))).)))).((((((((((.....))...)))))))).)))..))))).))).................................................. .........................................................................................90............................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | DRR000555(DRX000313) "THP-1 whole cell RNA, no treatment". (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR553575(SRX182781) source: Kidney. (Kidney) | DRR000556(DRX000314) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | DRR000558(DRX000316) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR326281(GSM769511) "Dicer mRNA was knocked down using siDicer, c. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR001488(DRX001042) "Hela short nuclear cell fraction, LNA(+)". (hela) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330857(SRX091695) tissue: skin psoriatic involveddisease state:. (skin) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR330910(SRX091748) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR038858(GSM458541) MEL202. (cell line) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553576(SRX182782) source: Testis. (testes) | SRR038857(GSM458540) D20. (cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR326282(GSM769512) "Dicer mRNA was knocked down using siDicer, t. (cell line) | TAX577738(Rovira) total RNA. (breast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR038862(GSM458545) MM472. (cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | TAX577580(Rovira) total RNA. (breast) | SRR207117(GSM721079) Whole cell RNA. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191397(GSM715507) 30genomic small RNA (size selected RNA from t. (breast) | TAX577588(Rovira) total RNA. (breast) | SRR037935(GSM510473) 293cand3. (cell line) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR207111(GSM721073) Whole cell RNA. (cell line) | DRR000557(DRX000315) "THP-1 whole cell RNA, after 3 day treatment . (cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR033725(GSM497070) Unmutated CLL (CLLU626). (B cell) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | TAX577741(Rovira) total RNA. (breast) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | TAX577590(Rovira) total RNA. (breast) | SRR038861(GSM458544) MM466. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | GSM956925Ago2D5(GSM956925) "cell line: HEK293 cell linepassages: 15-20ip. (ago2 cell line) | DRR001489(DRX001043) "Hela short nuclear cell fraction, control". (hela) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | TAX577589(Rovira) total RNA. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR038853(GSM458536) MELB. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR038856(GSM458539) D11. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | TAX577745(Rovira) total RNA. (breast) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR553574(SRX182780) source: Heart. (Heart) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................ATCCCTGTCCTGCCTTATG............................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................ATTGTCCCTTGCTATCCCTTATA...................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................AGGGATAGCAAGGGACAA.......................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ...........................................................................................................................................................................GTCCTGGTCAGTCTGG............................................................... | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - |
| ...........................................................................................................................................................................................................AGCTGCAGACAGTGGC............................... | 16 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - |