| (5) B-CELL | (1) BRAIN | (2) BREAST | (11) CELL-LINE | (4) HEART | (2) LIVER | (1) OTHER | (3) SKIN | (1) TESTES | (1) XRN.ip |
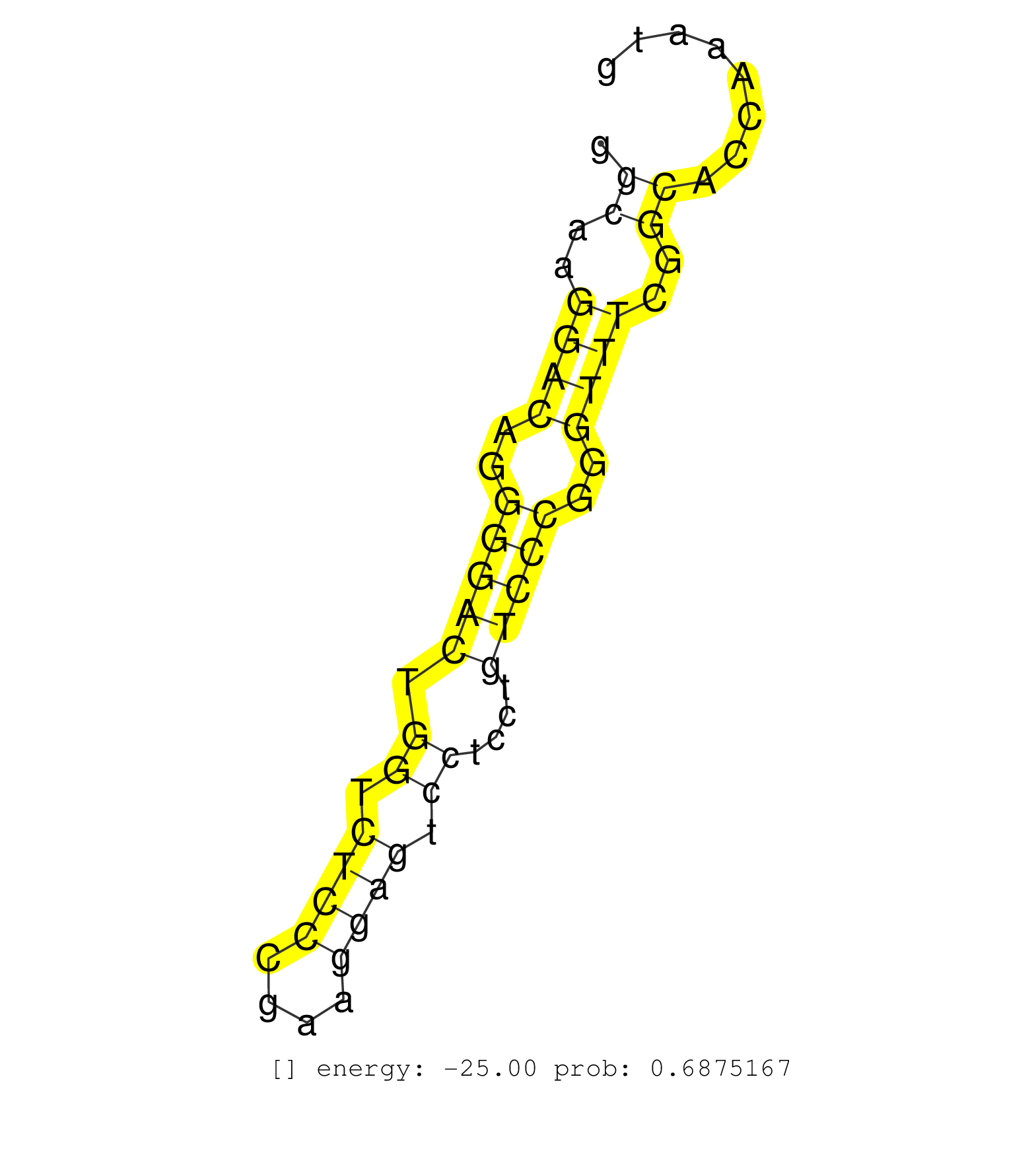
| TGCTGGTTAATTTTGCCACATATCACCTTGTGATGTTTACGGTAGCAGCTAGATGAACAGCAGGAACTTACAGAAGGGTGTAGGTAAAGTTTGTTTATGCTTCCCACGATCTCCCCTTGTGCGGTCCAGATGGTTTGTAATTGGTGAGGTCTGTTGGCTCTGCCCTAACACTGACCACAGTGCCCCTTCTGCCCTTCCAGTCTGCAGCCCCTCTTTCCTGGAGTAAATGAACTGGACCAAATCTCAAAAA ............................................................(((..(((((((......))))))).............(((...((.((((((.((...(((((.((....)).)))))...)).))))))))..))).)))........................................................................................ ............................................................61....................................................................................................163..................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR191611(GSM715721) 57genomic small RNA (size selected RNA from t. (breast) | SRR189782 | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................TGGTGAGGTCTGTTGTTT........................................................................................... | 18 | 24.00 | 0.00 | 7.00 | 2.00 | 2.00 | 1.00 | 2.00 | 2.00 | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | |
| .............................................................................................................................................TGGTGAGGTCTGTTGTTTT.......................................................................................... | 19 | 21.00 | 0.00 | 3.00 | 6.00 | 2.00 | 3.00 | 1.00 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................................AATGAACTGGACCAACA........ | 17 | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................TCTGTTGGCTCTGCCCTAACACTGAC........................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGTGAGGTCTGTTGAATA.......................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................GAGGTCTGTTGGCTCTGCCCT.................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................AGCTAGATGAACAGCAGGAACTTAC................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................GAGTAAATGAACTGGACCAAATC....... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................TGGTGAGGTCTGTTGAATT.......................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................CGGTAGCAGCTAGATGAACAGCAGGA......................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................GGTGAGGTCTGTTGGTTA.......................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................CTGTTGGCTCTGCCCGC................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TGGTGAGGTCTGTTGCTTT.......................................................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................ACTTACAGAAGGGTGTAGA...................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................TAGATGAACAGCAGGA......................................................................................................................................................................................... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |
| ....................................................................................................................................................GTCTGTTGGCTCTGCC...................................................................................... | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGCTGGTTAATTTTGCCACATATCACCTTGTGATGTTTACGGTAGCAGCTAGATGAACAGCAGGAACTTACAGAAGGGTGTAGGTAAAGTTTGTTTATGCTTCCCACGATCTCCCCTTGTGCGGTCCAGATGGTTTGTAATTGGTGAGGTCTGTTGGCTCTGCCCTAACACTGACCACAGTGCCCCTTCTGCCCTTCCAGTCTGCAGCCCCTCTTTCCTGGAGTAAATGAACTGGACCAAATCTCAAAAA ............................................................(((..(((((((......))))))).............(((...((.((((((.((...(((((.((....)).)))))...)).))))))))..))).)))........................................................................................ ............................................................61....................................................................................................163..................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR037937(GSM510475) 293cand2. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR191607(GSM715717) 192genomic small RNA (size selected RNA from . (breast) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR191611(GSM715721) 57genomic small RNA (size selected RNA from t. (breast) | SRR189782 | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................CACAAGGGGAGATCGTGGGAAGCAT................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TGCCCTTCCAGTCTGCAAGAT........................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................GCAGCCCCTCTTTCCTGCGC........................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................TGGTTTGTAATTGGTATCC..................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................CCAGTCTGCAGCCCCCTAG................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................TTCTGCCCTTCCAGTCTA.............................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................TACCGTAAACATCACAAGGTGATATGTG.............................................................................................................................................................................................................. | 28 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| ...................................................................................................................................................................GTGGTCAGTGTTAGG........................................................................ | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |