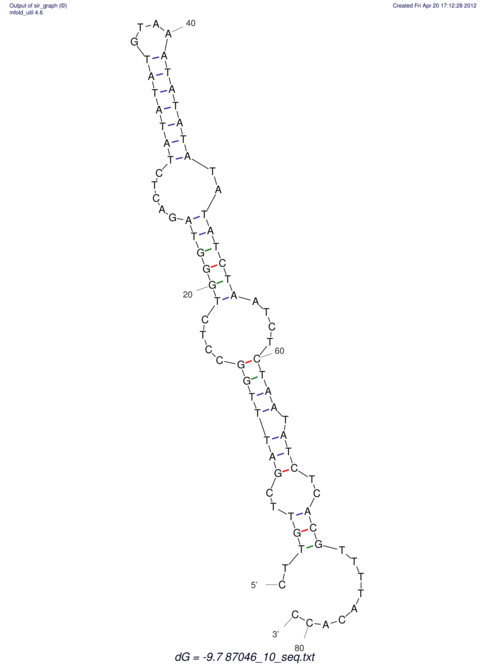
| TATGAAGTTAAAAAACGAAATCATGCTAGATCAAGAACAAATATCATGTAAATTG----CTTTTTTACA--TAAAATACAGGATATTCGTTCT-----------------------------TGAGCTTAATGGTTATAGAGC--------TGCAAGAAAAATTGT | Size | Hit Count | Total Norm | V113
Male body | V114
Embryo | V115
Head |
| .........................................................................AAATACAGGATATTCGTTCT-----------------------------T........................................... | 21 | 1 | 170.00 | 119.00 | 20.00 | 31.00 |
| .........................................................................AAATACAGGATATTCGTTCT-----------------------------TG.......................................... | 22 | 1 | 129.00 | 115.00 | 9.00 | 5.00 |
| ..........................................................................AATACAGGATATTCGTTCT-----------------------------TGA......................................... | 22 | 1 | 58.00 | 50.00 | 5.00 | 3.00 |
| ..........................................................................AATACAGGATATTCGTTCT-----------------------------TG.......................................... | 21 | 1 | 58.00 | 52.00 | 3.00 | 3.00 |
| .........................................................................AAATACAGGATATTCGTTCT-----------------------------TGT......................................... | 23 | 1 | 27.00 | 23.00 | 0.00 | 4.00 |
| ............................................................................TACAGGATATTCGTTCT-----------------------------TGAGC....................................... | 22 | 1 | 22.00 | 8.00 | 11.00 | 3.00 |
| ..........................................................................AATACAGGATATTCGTTCT-----------------------------TGT......................................... | 22 | 1 | 14.00 | 12.00 | 1.00 | 1.00 |
| .........................................................................AAATACAGGATATTCGTTCT-----------------------------............................................ | 20 | 1 | 11.00 | 6.00 | 0.00 | 5.00 |
| ..........................................................................AATACAGGATATTCGTTCT-----------------------------T........................................... | 20 | 1 | 9.00 | 6.00 | 0.00 | 3.00 |
| ..........................................................................AATACAGGATATTCGTTCT-----------------------------TGAA........................................ | 23 | 1 | 8.00 | 6.00 | 1.00 | 1.00 |
| .........................................................................AAATACAGGATATTCGTTCT-----------------------------TT.......................................... | 22 | 1 | 7.00 | 7.00 | 0.00 | 0.00 |
| ..........................................................................AATACAGGATATTCGTTCT-----------------------------TGAT........................................ | 23 | 1 | 5.00 | 5.00 | 0.00 | 0.00 |
| .........................................................................AAATACAGGATATTCGTTCT-----------------------------TGTA........................................ | 24 | 1 | 4.00 | 4.00 | 0.00 | 0.00 |
| ............................................................................TACAGGATATTCGTTCT-----------------------------TGAG........................................ | 21 | 1 | 3.00 | 2.00 | 0.00 | 1.00 |
| .........................................................................AAATACAGGATATTCGTTCT-----------------------------TGA......................................... | 23 | 1 | 3.00 | 3.00 | 0.00 | 0.00 |
| ............................................................................TACAGGATATTCGTTCT-----------------------------TGAGTT...................................... | 23 | 1 | 2.00 | 1.00 | 0.00 | 1.00 |
| ......................................................................................................................................GTAxGGAGC--------TGCAAGAAAA..... | 18 | 1 | 2.00 | 0.00 | 2.00 | 0.00 |
| ..........................................................................AATACAGGATATTCGTTCT-----------------------------TGAG........................................ | 23 | 1 | 2.00 | 2.00 | 0.00 | 0.00 |
| ..........................................................................AATACAGGATATTCGTTCT-----------------------------TGAAA....................................... | 24 | 1 | 2.00 | 1.00 | 0.00 | 1.00 |
| ............................................................................TACAGGATATTCGTTCT-----------------------------TGAGTTT..................................... | 24 | 1 | 2.00 | 2.00 | 0.00 | 0.00 |
| .........................................................................AAATACAGGACATTCGTTCT-----------------------------TG.......................................... | 22 | 1 | 2.00 | 2.00 | 0.00 | 0.00 |
| .........................................................................AGATACAGGATATTCGTTCT-----------------------------T........................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................AAATACAGGATATTCGTTCT-----------------------------TGAT........................................ | 24 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................AAATCCAGGATATTCGTTCT-----------------------------T........................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ................................................................TTACA--TAAAATAAAGTAT.................................................................................. | 18 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ..........................................................................AATACAGGATATTCGCTCT-----------------------------TGA......................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................................................................................TAGAGC--------TGCCAGAAAAGT... | 18 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .............................................................TCTTTACA--TAAAATACAGGATATTCG............................................................................. | 26 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ............................................................................TACAGGATATTCGTTCT-----------------------------TGAGTA...................................... | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................AAATACAGGATATTCGTTCC-----------------------------TGT......................................... | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ..........................................................................ACTACAGGATATTCGTTCT-----------------------------CG.......................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................AAATACAGGATATTCGCTGT-----------------------------TG.......................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................AAATACAGGATATTCGTTTT-----------------------------TG.......................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ..........................................................................AATACAGGATATTCGTTA.......................................................................... | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| .........................................................................AAATACAGGATATTCGTTGT-----------------------------TG.......................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................AAATACAGGATATTCGTTAT-----------------------------T........................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ............................................................................TACAGGATATTCGTTCT-----------------------------TGAGCTTTT................................... | 26 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................AAATACAGGATCTTCGTTCT-----------------------------TG.......................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ..........................................................................................................................................AGAGC--------TGGAAGAAAAATC.. | 18 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ............................................................................TACAGGATATTCGTTCT-----------------------------TGAGT....................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ..........................................................................AATACAGGATATTCGTTCG-----------------------------TGA......................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ............................................................................TACAGGATATTCGTTCT-----------------------------TGAGTTTT.................................... | 25 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ..........................................................................AATACGGGATATTCGTTCT-----------------------------TG.......................................... | 21 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .........................................................................AAATACAGGATATTCGTTCT-----------------------------TGAA........................................ | 24 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ......................ATGCTxGATGAAGAACAAA............................................................................................................................. | 18 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ...................................................................CA--TAAAATAAAGTATATT............................................................................... | 18 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .........................................................................AAATACAGGATATTCGTTAT-----------------------------TGT......................................... | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ............................................................................TACTGGATATTCGTTCT-----------------------------TGAGC....................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................AAATACAGGATATCCGTTCT-----------------------------TGT......................................... | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................AAATACAGGATATTCGTTC.......................................................................... | 19 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| .........................................................................AAATACAGGATATTCGTTGT-----------------------------T........................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................AAATACAGGATATTCGTTCT-----------------------------TGAAT....................................... | 25 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................AAATACAGGATATTCGCTGT-----------------------------T........................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ............................................................................TACAGGATATTCGTTCT-----------------------------TGAGTAT..................................... | 24 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................AAATxCAGGATATTCGTTCT-----------------------------TGAA........................................ | 23 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ...........................................................................ATACAGGATATTCGTTCT-----------------------------T........................................... | 19 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ..........................................................................AATACAGGATATTCGTTGT-----------------------------TGA......................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ..........................................................................AATACAGGATATTCGTTCT-----------------------------TGAGC....................................... | 24 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ............................................................................TACAGGATATTCGTTCT-----------------------------TGAGCTA..................................... | 24 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ............................................................................TACAGGATATTCGTTCT-----------------------------TGAGCTT..................................... | 24 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ..........................................................................AATACAGGATATTCGTTCT-----------------------------TAA......................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |