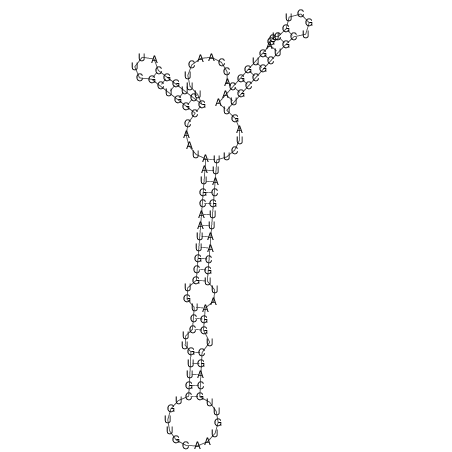
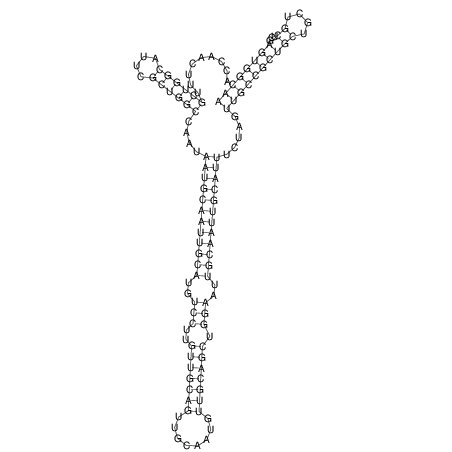
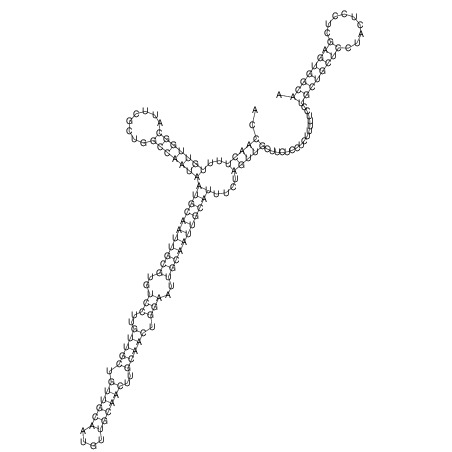
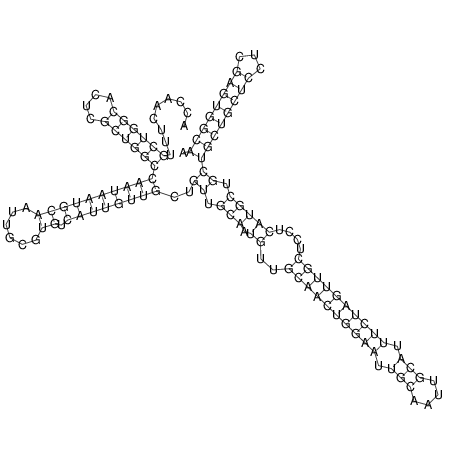
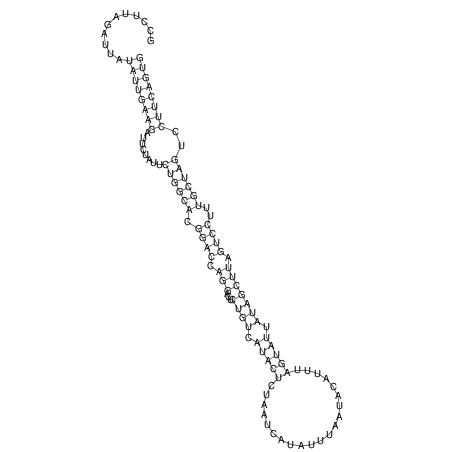
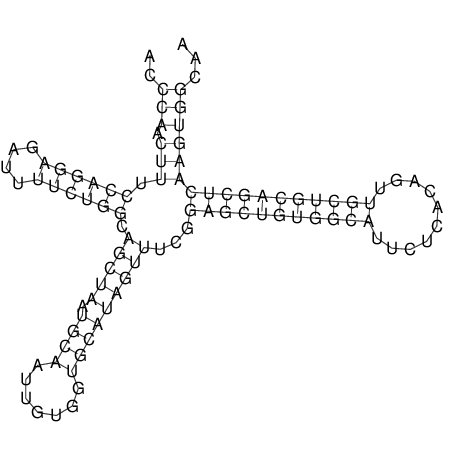
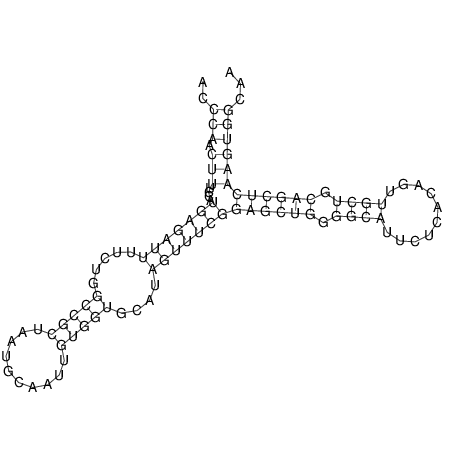
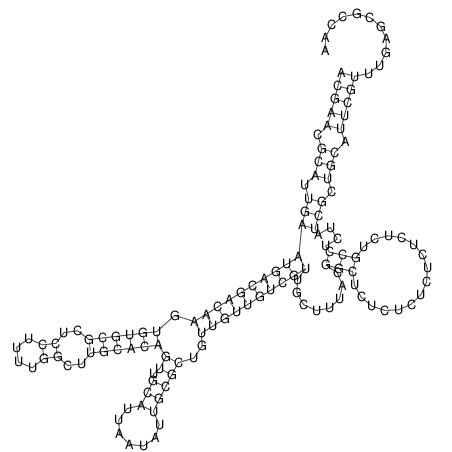
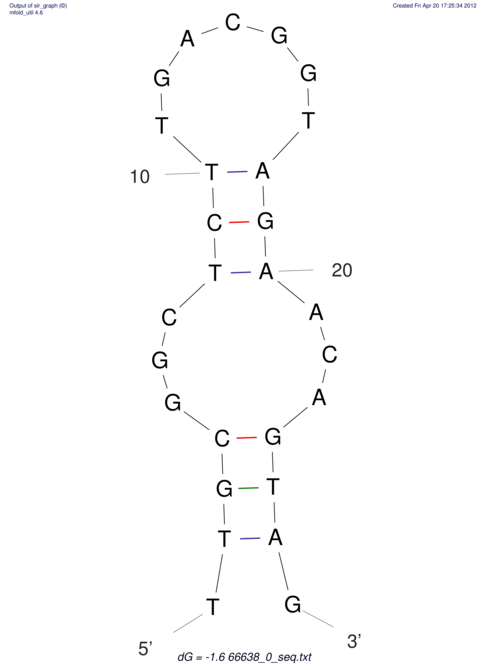
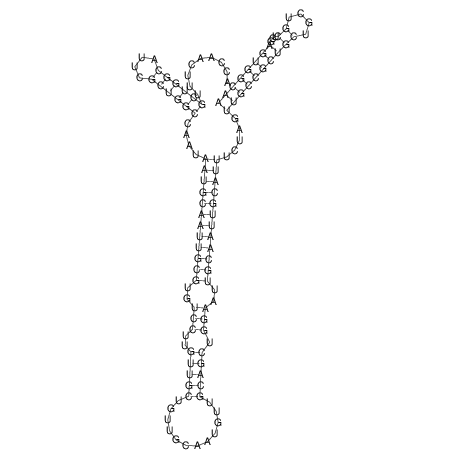
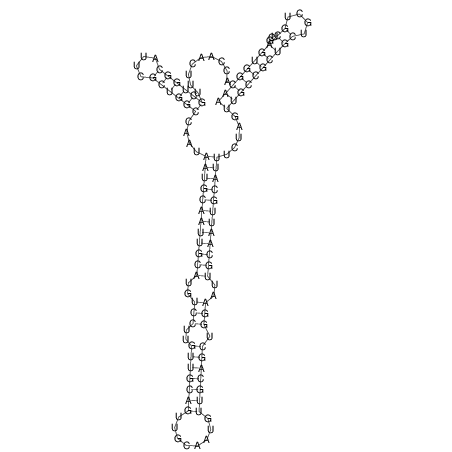
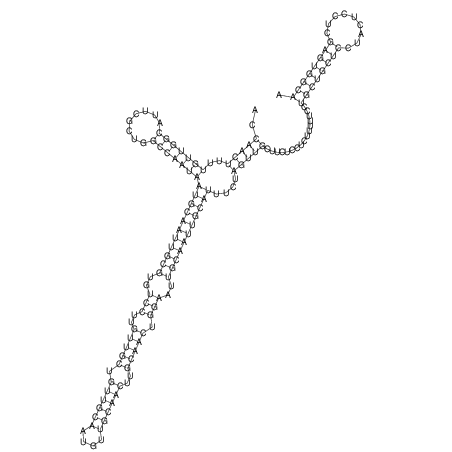
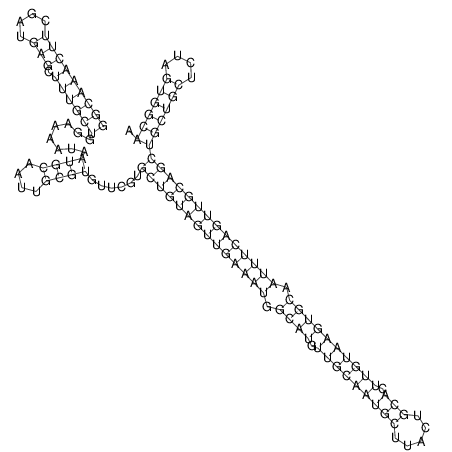
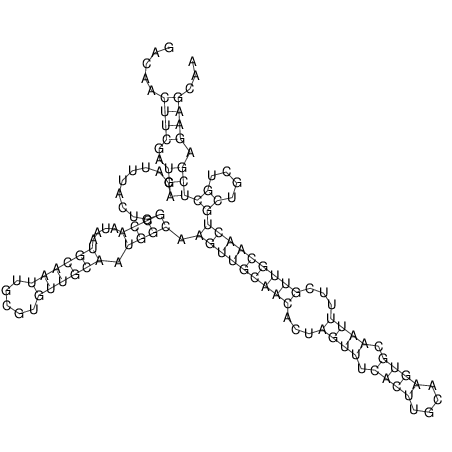
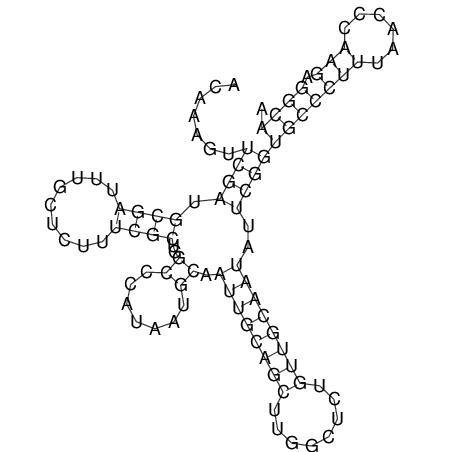
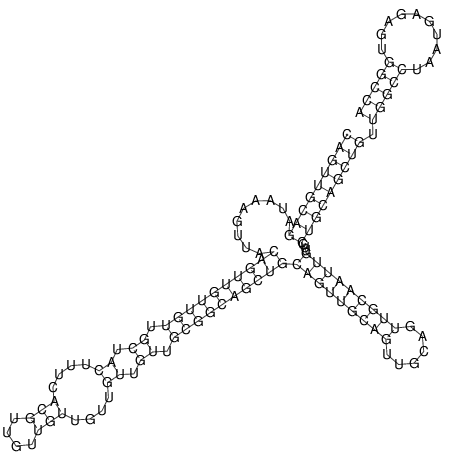
| TTCTGA--AAGG-GAAGAGCCTTAGATT-ATATTGAAGATTCTATTCTGGCACGGACCAGGACTCTGTCATACTCT-AATC----------------ATATTTAAT-----ACATTTAGTATTATAGCTT----------------------AGT---------------------------------------------------------------------------C--------------------C---TTTGCTAGTCCTTCAGTG---------------------------------------- | Size | Hit Count | Total Norm | V105
Male body | V106
Head | V055
Unknown | V039
Embryo |
| ...TGA--AAGG-GAAGGGCATT.................................................................................................................................................................................................................................................................................................. | 17 | 1 | 12.00 | 0.00 | 0.00 | 12.00 | 0.00 |
| .....A--AAGG-GAAGGGCCTTTG................................................................................................................................................................................................................................................................................................ | 17 | 1 | 8.00 | 0.00 | 0.00 | 8.00 | 0.00 |
| ....GA--AAGG-GAAGAGCATTTGA............................................................................................................................................................................................................................................................................................... | 19 | 1 | 7.00 | 7.00 | 0.00 | 0.00 | 0.00 |
| ....GA--AAGG-GAAGAGCATTTGAT.............................................................................................................................................................................................................................................................................................. | 20 | 1 | 3.00 | 3.00 | 0.00 | 0.00 | 0.00 |
| ....GA--AAGG-GAAGGGCCTTTGAT.............................................................................................................................................................................................................................................................................................. | 20 | 1 | 3.00 | 3.00 | 0.00 | 0.00 | 0.00 |
| ...TGA--AAGG-GAAGGGCAT................................................................................................................................................................................................................................................................................................... | 16 | 1 | 3.00 | 0.00 | 0.00 | 3.00 | 0.00 |
| ...TGA--AAGG-GAAGAGCATTTG................................................................................................................................................................................................................................................................................................ | 19 | 1 | 3.00 | 3.00 | 0.00 | 0.00 | 0.00 |
| ....GA--AAGG-GAAGGGCATT.................................................................................................................................................................................................................................................................................................. | 16 | 1 | 3.00 | 0.00 | 0.00 | 3.00 | 0.00 |
| .....A--AAGG-GAAGGGCCTTTGA............................................................................................................................................................................................................................................................................................... | 18 | 1 | 3.00 | 3.00 | 0.00 | 0.00 | 0.00 |
| .....A--AAGG-GAAGAGCATTTGA............................................................................................................................................................................................................................................................................................... | 18 | 1 | 2.00 | 1.00 | 0.00 | 1.00 | 0.00 |
| .....A--AAGG-GAAGGGCATTAG................................................................................................................................................................................................................................................................................................ | 17 | 1 | 2.00 | 0.00 | 0.00 | 2.00 | 0.00 |
| ...TGA--AAGG-GAAGAGCATTTGAT.............................................................................................................................................................................................................................................................................................. | 21 | 1 | 2.00 | 2.00 | 0.00 | 0.00 | 0.00 |
| .....A--AAGG-GAAGGGCATTAGA............................................................................................................................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | 0.00 | 0.00 | 0.00 |
| ....GA--AAGG-GAAGAGCATTTG................................................................................................................................................................................................................................................................................................ | 18 | 1 | 2.00 | 2.00 | 0.00 | 0.00 | 0.00 |
| ...TGA--AAGG-GAAGAGCATTT................................................................................................................................................................................................................................................................................................. | 18 | 1 | 2.00 | 2.00 | 0.00 | 0.00 | 0.00 |
| ....GA--AAGG-GAAGAGCATTT................................................................................................................................................................................................................................................................................................. | 17 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ....GA--AAGG-GAAGGGCGTTAG................................................................................................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ........AAGG-GAAGGGCATTAGAT.............................................................................................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| .....A--AAGG-GAAGGGCATTAGAT.............................................................................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....A--AAGG-GAAGAGCATTAGA............................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ...TGA--AAGG-GAAGAGCATTTGA............................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ...TGA--AAGG-GAAGGGCCTTT................................................................................................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| .....A--AAGG-GAAGCGCCTTTG................................................................................................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| .....A--AAGG-GAAGAGCATTTG................................................................................................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| .................................................................................................ATACTTCAT-----ACATTTAGT................................................................................................................................................................................................. | 18 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....GA--AAGG-GAAGGGCATTAGAT.............................................................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ..TTGA--AAGG-GAAGGGCCTT.................................................................................................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ....................CTTGGATT-xTATTGAAGA.................................................................................................................................................................................................................................................................................. | 17 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ..............................................CTGGCACGGACTTGGA........................................................................................................................................................................................................................................................... | 16 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ...TGA--AAGG-GAAGCGCAT................................................................................................................................................................................................................................................................................................... | 16 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| .....A--AAGG-GAAGGGCCTTAGATC-............................................................................................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ........AAGG-GAAGGGCCTTTGA............................................................................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ....GA--AAGG-GAAGGGCCTTTG................................................................................................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ........AAGG-GAAGCGCCTTTGA............................................................................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |