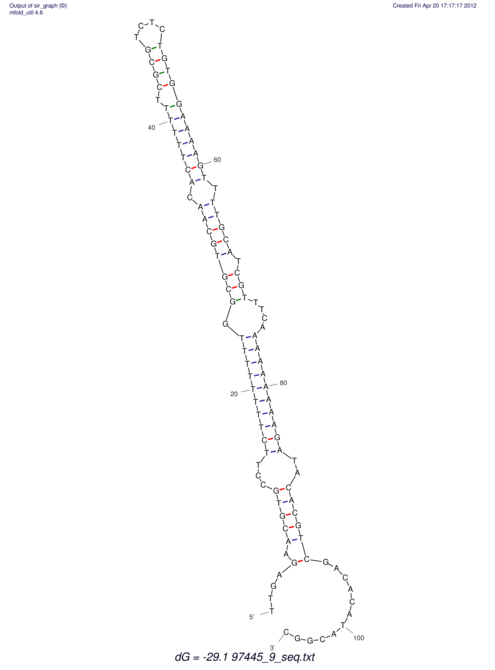
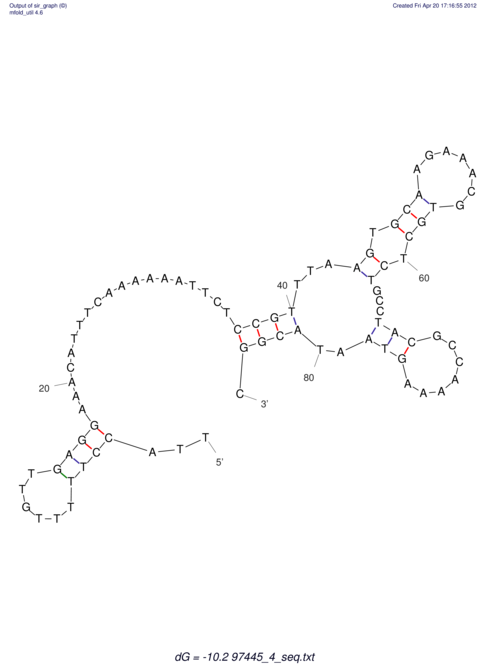
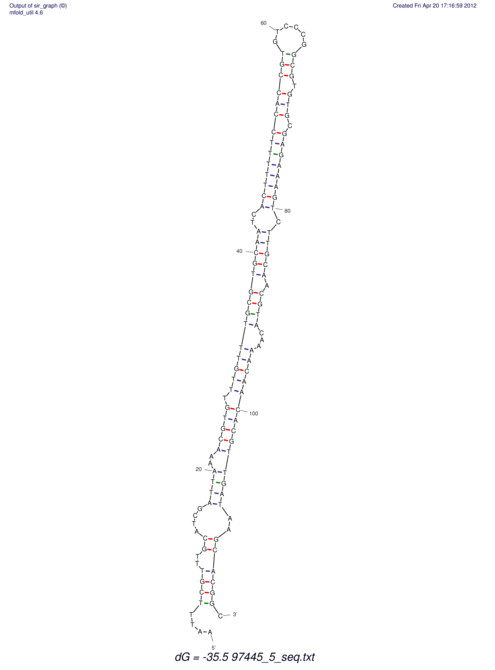
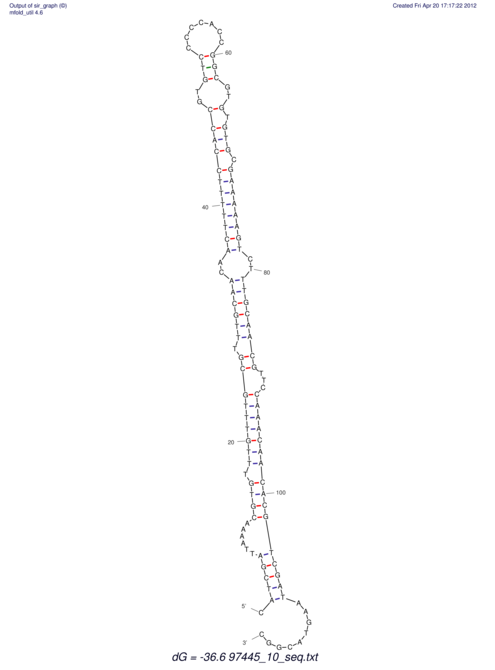
| dm3 |
chr2R:3294222-3294357 - |
|
1 |
TTTTCGTTTACTTT-TTA---ACC-------CTCCGCT--CGAGAACGTGGTTTCT---TTTGCGA--CGTGCAATAC--TC----------------------------------------TGTGGCAGAAGTC-TTGCACCGTTC-----------------CAAAAGA-GAGC-AGCACGTGGAGCGACGAGAACGGCCGTA-CAATACCCAAG |
| droSim1 |
chrU:460991-461115 - |
LASTZ |
1 |
TTTT------------TA---ACC-------CTCCGCT--CGAGAACGTGGTTTCT---TTTGCGA--CGTGCAATAC--TC----------------------------------------TGTGGTTCAAGTC-TTGCACCGTTC-----------------CAAAAGA-GAGC-AACACGTGGAGCGACGAGCACGGCCGTA-CAATACCCAAG |
| droSec1 |
super_1:944262-944397 - |
LASTZ |
1 |
TTTTCGTTTACTTT-TTA---ACC-------CTCCGCT--CGAGAACGTGGTTTCT---TTTGCGA--CGTGCAATAC--TC----------------------------------------TGTGGCAGAAGTC-TTGCACCGTTC-----------------CAAAAGA-GAGC-AGCACGTGGAGCGACGAGCACGGCCGTA-CAATACCCAAG |
| droYak2 |
chr2L:15977521-15977656 - |
LASTZ |
1 |
TTTTCGTTTACTTG-TTA---ACC-------CTCCGCT--CGAGAACGTGGTTTCT---TTTGCGA--CGTGCAATAC--TC----------------------------------------TGTGGCAGAAGTC-TTGCATCGTTC-----------------CAAAAGA-GAGC-AGCACGTGGAGCGACGAGCACGGCCGTA-AAATACCCAAG |
| droEre2 |
scaffold_4929:19300554-19300689 + |
LASTZ |
4 |
TTTTCGTTTACTTT-TTA---ACC-------CTCCGCT--CGAGAACGTGGTTTCT---TTTGCGA--CGTGCAATAC--TT----------------------------------------TGTGGCAGAAGTC-TTGCACCGTTC-----------------CAAAAGA-GAAC-AGCACGTGGAGCGACGAGCACGGCCGTA-CAATACCCAAG |
| droEug1 |
scf7180000409672:5307008-5307144 - |
LASTZ |
4 |
TTTTCGTTTACTTT-TTA---ACC-------CTCCGCT--CGAGAACGTGGTTTCT---TTTGCGA--CGTGCAATAC--TT----------------------------------------TGTGGCAAAAGTC-TTGCATCGTTC-----------------CAAAAGA-GAGCAACCACTTGGAGCGACGAGCACGGCCGTA-CAATACCCAAA |
| droBia1 |
scf7180000302292:2853030-2853170 + |
LASTZ |
4 |
TTTTGGTTTAATTT-TTGAACACC-------CTCCGCT--CGAGAACGTGGTTTCT---TTTGCGACGCGTGCAATAC--TT----------------------------------------TGTGGCAGAAGTC-TTGCATCGTTC-----------------CAAAAGA-GAGC-AGCACGTGGAGCGACGAGCACGGCCGTA-CAATACCCAAG |
| droTak1 |
scf7180000412807:59993-60132 - |
LASTZ |
4 |
TTTTCGTTTAATTT-TCC-ACACC-------CTCCGCT--CGAGAACGTGGTTTCT---TTTGCGACGCGTGCAATAC--TT----------------------------------------TGTGGCAAAAGTC-TTGCATCGTTC-----------------CAAAAGA-GAGC-AGCACGTGGAGCGACGAGCACGGCCGTA-AAATACCCAAA |
| droEle1 |
scf7180000491347:110639-110775 - |
LASTZ |
4 |
CTTTCGTTTACTTT-TTG---ACC-------CTCCGCT--CGGGAACGTGGTTTCT---TTTGCGA--CGTGCAATAC--TT----------------------------------------TGTGGCCAAAGTC-TTGCAACGTTC-----------------CAAAGGA-GAGC-TGCACGTGGAGCGACGAGTACGGCCGTAAAAATACCCAAG |
| droRho1 |
scf7180000770698:29092-29228 - |
LASTZ |
4 |
TTTTCATTTACTTT-TTA---ACC-------CTCCGCT--CGAGAACGTGGTTTCT---TTTGCGA--CGTGCAATAC--TT----------------------------------------TGTGGCAAAAGTT-TTGCAACGTTC-----------------CAAAGGA-GAGC-AGCACGTGGAGCGACGAGTACGGCCGTAAAAATACCCAAG |
| droFic1 |
scf7180000453948:50042-50182 - |
LASTZ |
4 |
TTTTCGTTTACTTTCTTA--AACC-------CTCCGCT--CGAGAACGTGGTTTCT---TTTGCGA--CGTGCAATTC--TTT---------------------------------------TGTGGCAAAAGTC-TTGCATCGTTC-----------------CAAAAGACTAAC-AGCACGTGGAGCGACAAGTACGGCCGTAAAAATACCCAAA |
| droKik1 |
scf7180000302670:19296-19422 - |
LASTZ |
4 |
TTTTCGTT-AGTTT--------CC-------CTCCGTT--CGCGAGCGTGTTTCCT----TTG-GG--CGTGCAACAC--TT----------------------------------------TTTGCCAAAAGTC-TTGCCACTGTC-----------------CAAAATC-GCGA-AGCACGTCGA---ACGAGTACGGCCGTAAAAATTCCCAAG |
| droAna3 |
scaffold_13266:3498798-3498932 - |
LASTZ |
4 |
ACCCC-TTTATTTT-TAT---TTT-------CACCGTT--CGAGAGGGTGTTCTCTTTGTTTTCGG--CGTGCAAAAC--CC----------------------------------------TTTG--AGAAGTT-TTGCATCGTTC-----------------GAAATTAGGGGC-AGCACCTCGAACA--GTACACGGCCGTA-AAATTCCTAAA |
| droBip1 |
scf7180000396427:1965389-1965535 - |
LASTZ |
4 |
ACCCC-TTTATTTT-TAT---TTT-------CACCGTT--CGAGAGGGTGTTCTCTTAGTTTTCGG--CGTGCAACAC--TTTTTTC----------------------------GCGTCCGTGTGG-AAAAGTCGTTGCATCGTTC-----------------GAA--CT-GGAC-AGCACCTCGAACAGC--ATACGGCCGCAAAAATTCCTAAA |
| dp4 |
chr3:1570003-1570137 - |
LASTZ |
4 |
TTTTTGGTTTTGTT-----------------------T--TGAGAACGTGCCTTCTTT-TTTTTGG--CGTGCAACAC--TTTTTTC----------------------------GCGTCTCTGTGG-AAAAGTT-TTGCATCGTTT-----------------CAAAAAAAAAGA-TACACGTCG----ACACATACGGCCGCTAAGATTCCAAAA |
| droPer1 |
super_2:1744183-1744317 - |
LASTZ |
4 |
TTTTTGGTTTTGTT-----------------------T--TGAGAACGTGCCTTCTTT-TTTTTGG--CGTGCAACAC--TTTTTTC----------------------------GCGTCTCTGTGG-AAAAGTT-TTGCATCGTTT-----------------CAAAAAAAAAGA-TACACGTCG----ACACATACGGCCGCTAAGATTCCAAAA |
| droWil1 |
Unknown |
MAF |
0 |
TTTTTTTTTTTTGT-TTT---ACC-------TTTTTGT--TGAGGAA---------------------------ACAT--TT------------------------------------------CA--AAAAATT-CT---CCGTTTAAGTGCAAGAAACGTGCTCTG------CC---TACGCCA--AAAGTAATACGGCCGTAAACATTCCAAAA |
| droVir3 |
scaffold_12875:10810504-10810650 - |
LASTZ |
4 |
TTTTGATTTACTTT---A---ATTTCGTTTGCATCGAT--T-AAAACGTGTT-TG----TTTG-----CGTGCAATCA-CTTTTTCC-ACCGTGTC----------------CCGGC--GTGTGCG-AGAAAGTC-TTGCAACGTAC-----------------AAA-------AC-AACACGTTG----ATAAGCACGGCCGCAAAAATTCCCAAA |
| droMoj3 |
scaffold_6496:16132312-16132448 + |
LASTZ |
4 |
TTTTG--------------------------CATCGAT--T-AAAACGTGTTT--G---TTTGCG---TTTGCAACAA-CTT-TTTCCACCGTGTCCCCCAC-----------CGGCGTGTGTGCG-AAAAAGTCTTTGCAACGTTC-----------------CAAA-------C-AACACGTCG----ATAAGTACGGCCGCAAAAATTCCTAAA |
| droGri2 |
scaffold_15245:3916304-3916452 - |
LASTZ |
4 |
TTTTCGTTTGC---------------------ATCGATTGT-GAAACGTGTTT--G---TTTGTAG--CGTGCAACACACTTTTT-C-AC----T--CCCACCCAGCGTGTGT--GTGTGTGTGTG-AAAAAGTCTGTGCAACGTTC-----------------AGAA------AC-AACACGTCGAAC----TCTACGGCCGCAAAAATTCCCAAA |