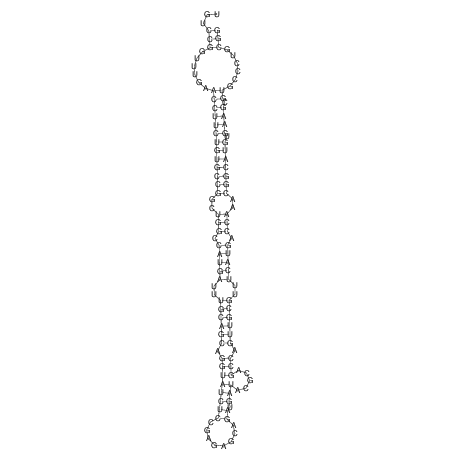
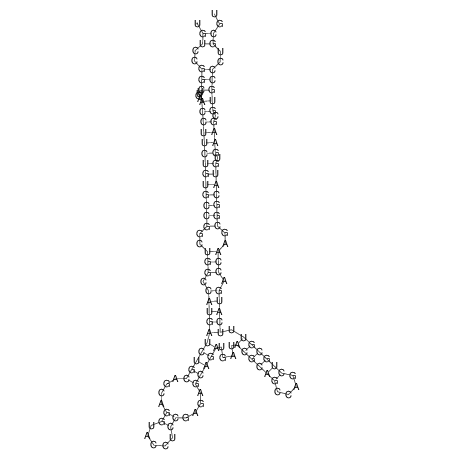

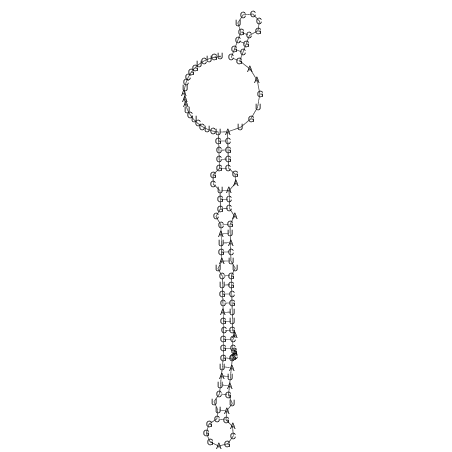
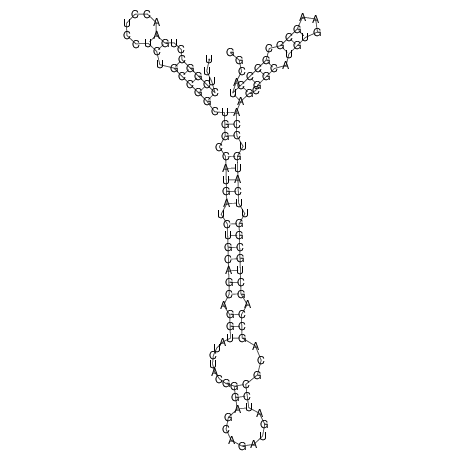


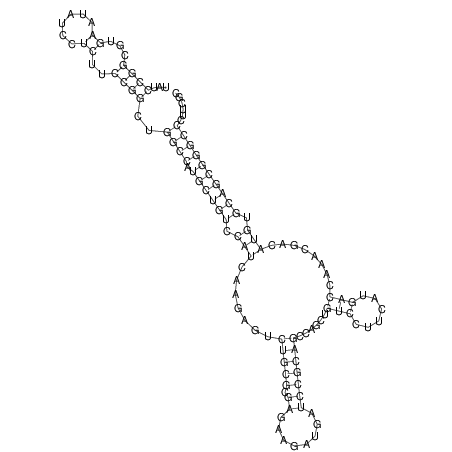
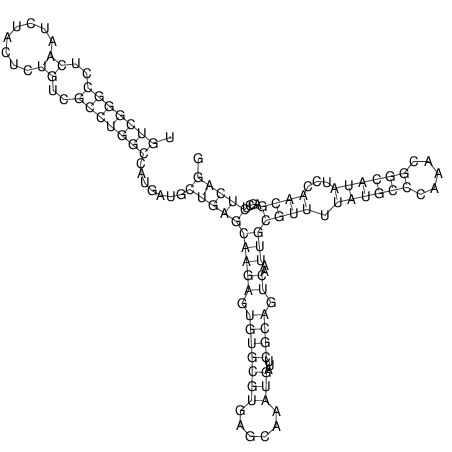
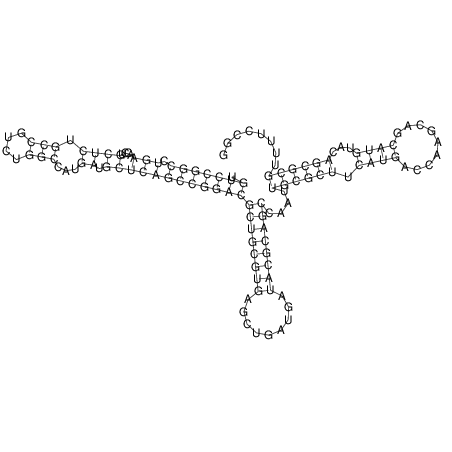

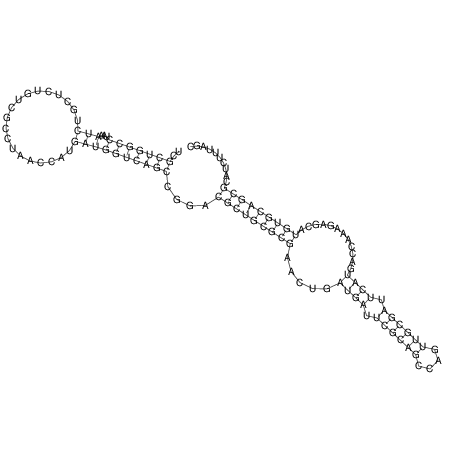



View alignment of MAF (12-files) vs LASTZ top hits (20-flies)
View all possible LASTZ hits (20-flies)
dme-mir-4959 |
chrX:19063163-19063281 - |
Known_mirbase |
|---|---|---|
| [View on miRBase] | [View on UCSC Genome Browser {Cornell Mirror}] | CDS |
Legend:
mature strand at position 19063234-19063255Notes: "As with all CDS miRNAs, the structure of this miRNA varies widely from one species to another. "
| Species | Coordinate | Source | Confidence | Alignment |
| dm3 | chrX:19063148-19063296 - | 1 | TGCTCATGGCCCTGATGTCCGGTTTGAACCTTCTGTGCCGGCTGGCCATGATTTGCAGCAGGTATCTCCGAGAGCAGATGATACGCAGCCAGTTGCGTTTCATGACCAAACGGCATGTGAAGCGTGCCCTGCGGGACCTGACCATCGGC | |
| droSim1 | chrX:14724057-14724205 - | LASTZ | 4 | TGCTCATGGCCCTGATGTCCGGCTTGAACCTTCTGTGCCGGCTGGCCATGATCTGCAGCAGGTACCTCCGAGAGCAGATGATACGCAGCCAGCTGCGTTTCATGACCAAGCGGCATGTGAAGCGTGCCCTGCGTGACCTGACCATCGGC |
| droSec1 | super_8:1352288-1352436 - | LASTZ | 1 | TGCTCATGGCCCTGATGTCCGGCTTGAACCTTCTGTGCCGGCTGGCCATGATCTGCAGCAGGTATCTCCGAGAGCAGATGATACGCAGCCAGCTGCGTTTCATGACCAAGCGGCATGTGAAGCGTGCCCTGCGTGACCTGACCATCGGC |
| droYak2 | chrX:17654811-17654959 - | LASTZ | 4 | TACTGATGGCCCTGATGTCTGGCCTAAATCTCCTCTGCCGGCTGGCCATGATCTGCAGCGGGTATCTTCGGGAGCAGATGATACGCAGCCAGTTGCGGTTCATGACCAAGCGGCATGTGAAGCGCGCCCTGCGCGACCTCACCATCGGC |
| droEre2 | scaffold_4690:9326382-9326530 - | LASTZ | 3 | TGCTGATGGCCCTGATTTCCGGCCTGAACCTCCTCTGCCGGCTGGCCATGATCTGCAGCAGGTATCTACGGGAGCAGATGATCCGCAGCCAGCTGCGGTTCATGTCCAAGCGGCATGTGAAGCGCGCCCTACGGGACCTCACCATCGGC |
| droEug1 | scf7180000409089:53868-54016 + | LASTZ | 5 | TGCTGATGGCTTTTATGTCTGGACTGAATCTTCTCTGCCGACTGGCCATGATCCTTAGCAGGAATCTCCGCGAGCAGATGATTCGCAGTCAGTTGCGTTTCATGAGCAAAAGGCATGTGAAGCGAGCCCTGGGCGACCTAACCATTGGG |
| droBia1 | scf7180000302187:38542-38690 + | LASTZ | 5 | TCCTGATGGCCCTGATGTCCGGCCTGAACCTCCTCTCCCGGCTGGCCATGATCTGCAGCAGATACCTCCGCGAGCAGATGATCCGCAGCCAGCTGCGCTTCATGACCAAGCGGCATGTGAAGCGCGCTCTGCGCGACCTCACCATCGGG |
| droTak1 | scf7180000415281:63891-64039 - | LASTZ | 5 | TCCTGATGGCCCTGATGTCCGGCCTGAATCTGCTCTGTCGGCTGGCGATGATCTGCAGCAGGAATCTGAGGGAGCAGATGATCCGCAGCCAGTTGCGCTTCATGGCCAAGCGGCATGTGAAGCGCGCCCTGCGGGATCTGACCATCGGG |
| droEle1 | scf7180000490751:647069-647217 - | LASTZ | 5 | TGCTGATGGCCCTGATGTCCGGCCTGAATCTCCTCTGCCGGCTGGCCATGATCTGCAGCAGGAAACTGCGGGAGCAGATGATCCGCAGTCAGCTGCGATTCATGACCAAGCGGCATGTGCAGCGCGCCCTGCGCGACCTCACCATTGGC |
| droRho1 | scf7180000778091:273360-273508 - | LASTZ | 5 | TGCTGATGGCCCTCATGTCCGGCCTGAATCTCCTTTGCCGGCTGGCGATGATCTGCAGCAGGAATCTGCGGGAGCAGATGATCCGCAGCCAGCTGCGCTTCATGACCAAGCGGCATGTGCAGCGGGCACTGCGCGACCTCACCATCGGC |
| droFic1 | scf7180000451789:82400-82548 - | LASTZ | 5 | TGCTGATGGCCCTAATGTCCGGACTGAATCTCCTCTGCCGGCTGGCGATGATCTGCAGCCGGAACTTGCGGGAGCAGATGATTCGCAGCCAGCTGCGCTTCATGACCAAGCGGCATGTGCAGCGCGCCCTGCGCGACTTAACCATCGGT |
| droKik1 | scf7180000302234:53857-54005 + | LASTZ | 5 | TGCTGATGGCCCTGATGTCCGGCCTGAATCTGCTCTGTCGGCTGGCCATGATCCTGAGCAGTTACCTGCGCGAACAGATGATTCGCAGCCAGTTGCGGTTCATGAGCATGCGGCATGTGCAGCGCGCTTTGCGGGACCTGACCATCGGT |
| droAna3 | scaffold_12903:667658-667806 + | LASTZ | 5 | TCCTGATGGCCCTGATGTCCGGCCTGAATTTGGTCTGTCGCGTGGCGATGATTATCAGCAAGACCCTAAGAGAGCAGATGATCCGGAGCCAGTTGAGGTTCATGAAGAAGCGGCATGTCCAGAGGGCGCTGAGGGATCTGACGATCGGG |
| droBip1 | scf7180000395916:40365-40513 - | LASTZ | 5 | TGCTCATGGCCCTGATGTCCGGTCTGAACTTGGTCTGTCGCCTGGTGATGATTATCAGTAAGACCTTACGGGAACAGATGATCCGAAGCCAGTTGAGGTTCATGAAGAAGCGGCATGTGCAGAGGGCGCTGAGGGATCTGACGATTGGG |
| dp4 | chrXL_group1e:10756223-10756371 - | LASTZ | 5 | TCCTGATAGCCATCATATCCGGCGTGAATATCCTCTTCCGGCTGGCCATGCTGTCCATCAAGAGTCTGCGCGAGAAGATGATCCGCAGCCAGCTGTCCTTCATGACCAAACGACATGTGCAGCGGGCCCTGCGCGACCTCACCATCGGG |
| droPer1 | super_14:190427-190575 - | LASTZ | 5 | TCCTGATAGCCATCATATCCGGCGTGAATATCCTCTTCCGGCTGGCCATGCTGTCCATCAAGAGTCTGCGCGAGAAGATGATCCGCAGCCAGCTGTCCTTCATGACCAAACGACATGTGCAGCGGGCCCTTCGCGACCTCACCATCGGG |
| droWil1 | scaffold_181096:8247011-8247159 - | LASTZ | 5 | TGCTGATGGCCATCATGTCGGGCCTCAATCTACTCTGTCGCCTGGCCATGATGCTGAGCAAGAGTGTGCGTGAGCAAATGATTCGCAGTCAATTGCGTTTTATGCCCAAACGGCATATCCAACGAGCTCTCAGGGATCTAACCATTGGC |
| droVir3 | scaffold_12726:998444-998592 - | LASTZ | 5 | TGCTGATGGCCTTCATGTCCGGCCTGAACCTGCTCTGCCGTCTGGCCATGATGCTCAGCCGGACGCTGCGTGAGCTGATGATACGCAGCCAATTGCGCTTCATGACCAAGCAGCATGTACAGCGCGTTTTCCGGGATCTAACCATCGGT |
| droMoj3 | scaffold_6473:9166585-9166733 + | LASTZ | 5 | TGCTGATGGCCTTCATGTCCGGCCTGAATCTGCTCTGTCGCCTGGCCATGATGCTCAGCCGGACGCTGCGCGAGCTGATGATACGCAGCCAATTGCGCTATATGACCAAACAGCATGTGCAGCACGTCTTCAGGGATCTGACCATCGGC |
| droGri2 | scaffold_14853:5433396-5433544 - | LASTZ | 5 | TCCTCATGGCGATCATCGCTGGCCTAAATCTGCTCTGTCGCCTAACCATGATGGTCAGCCGGACGCTGCGCGAACTGATGATTCGCAGCCAGTTGCGATTCATGACCAAAGAGCATGTGCAGCGCATCTTTAGCCATTTAACCATCGGC |
| Species | Alignment |
| dm3 | TGCTCATGGCCCTGATGTCCGGTTTGAACCTTCTGTGCCGGCTGGCCATGATTTGCAGCAGGTATCTCCGAGAGCAGATGATACGCAGCCAGTTGCGTTTCATGACCAAACGGCATGTGAAGCGTGCCCTGCGGGACCTGACCATCGGC |
| droSim1 | TGCTCATGGCCCTGATGTCCGGCTTGAACCTTCTGTGCCGGCTGGCCATGATCTGCAGCAGGTACCTCCGAGAGCAGATGATACGCAGCCAGCTGCGTTTCATGACCAAGCGGCATGTGAAGCGTGCCCTGCGTGACCTGACCATCGGC |
| droSec1 | TGCTCATGGCCCTGATGTCCGGCTTGAACCTTCTGTGCCGGCTGGCCATGATCTGCAGCAGGTATCTCCGAGAGCAGATGATACGCAGCCAGCTGCGTTTCATGACCAAGCGGCATGTGAAGCGTGCCCTGCGTGACCTGACCATCGGC |
| droYak2 | TACTGATGGCCCTGATGTCTGGCCTAAATCTCCTCTGCCGGCTGGCCATGATCTGCAGCGGGTATCTTCGGGAGCAGATGATACGCAGCCAGTTGCGGTTCATGACCAAGCGGCATGTGAAGCGCGCCCTGCGCGACCTCACCATCGGC |
| droEre2 | TGCTGATGGCCCTGATTTCCGGCCTGAACCTCCTCTGCCGGCTGGCCATGATCTGCAGCAGGTATCTACGGGAGCAGATGATCCGCAGCCAGCTGCGGTTCATGTCCAAGCGGCATGTGAAGCGCGCCCTACGGGACCTCACCATCGGC |
| droEug1 | TGCTGATGGCTTTTATGTCTGGACTGAATCTTCTCTGCCGACTGGCCATGATCCTTAGCAGGAATCTCCGCGAGCAGATGATTCGCAGTCAGTTGCGTTTCATGAGCAAAAGGCATGTGAAGCGAGCCCTGGGCGACCTAACCATTGGG |
| droBia1 | TCCTGATGGCCCTGATGTCCGGCCTGAACCTCCTCTCCCGGCTGGCCATGATCTGCAGCAGATACCTCCGCGAGCAGATGATCCGCAGCCAGCTGCGCTTCATGACCAAGCGGCATGTGAAGCGCGCTCTGCGCGACCTCACCATCGGG |
| droTak1 | TCCTGATGGCCCTGATGTCCGGCCTGAATCTGCTCTGTCGGCTGGCGATGATCTGCAGCAGGAATCTGAGGGAGCAGATGATCCGCAGCCAGTTGCGCTTCATGGCCAAGCGGCATGTGAAGCGCGCCCTGCGGGATCTGACCATCGGG |
| droEle1 | TGCTGATGGCCCTGATGTCCGGCCTGAATCTCCTCTGCCGGCTGGCCATGATCTGCAGCAGGAAACTGCGGGAGCAGATGATCCGCAGTCAGCTGCGATTCATGACCAAGCGGCATGTGCAGCGCGCCCTGCGCGACCTCACCATTGGC |
| droRho1 | TGCTGATGGCCCTCATGTCCGGCCTGAATCTCCTTTGCCGGCTGGCGATGATCTGCAGCAGGAATCTGCGGGAGCAGATGATCCGCAGCCAGCTGCGCTTCATGACCAAGCGGCATGTGCAGCGGGCACTGCGCGACCTCACCATCGGC |
| droFic1 | TGCTGATGGCCCTAATGTCCGGACTGAATCTCCTCTGCCGGCTGGCGATGATCTGCAGCCGGAACTTGCGGGAGCAGATGATTCGCAGCCAGCTGCGCTTCATGACCAAGCGGCATGTGCAGCGCGCCCTGCGCGACTTAACCATCGGT |
| droKik1 | TGCTGATGGCCCTGATGTCCGGCCTGAATCTGCTCTGTCGGCTGGCCATGATCCTGAGCAGTTACCTGCGCGAACAGATGATTCGCAGCCAGTTGCGGTTCATGAGCATGCGGCATGTGCAGCGCGCTTTGCGGGACCTGACCATCGGT |
| droAna3 | TCCTGATGGCCCTGATGTCCGGCCTGAATTTGGTCTGTCGCGTGGCGATGATTATCAGCAAGACCCTAAGAGAGCAGATGATCCGGAGCCAGTTGAGGTTCATGAAGAAGCGGCATGTCCAGAGGGCGCTGAGGGATCTGACGATCGGG |
| droBip1 | TGCTCATGGCCCTGATGTCCGGTCTGAACTTGGTCTGTCGCCTGGTGATGATTATCAGTAAGACCTTACGGGAACAGATGATCCGAAGCCAGTTGAGGTTCATGAAGAAGCGGCATGTGCAGAGGGCGCTGAGGGATCTGACGATTGGG |
| dp4 | TCCTGATAGCCATCATATCCGGCGTGAATATCCTCTTCCGGCTGGCCATGCTGTCCATCAAGAGTCTGCGCGAGAAGATGATCCGCAGCCAGCTGTCCTTCATGACCAAACGACATGTGCAGCGGGCCCTGCGCGACCTCACCATCGGG |
| droPer1 | TCCTGATAGCCATCATATCCGGCGTGAATATCCTCTTCCGGCTGGCCATGCTGTCCATCAAGAGTCTGCGCGAGAAGATGATCCGCAGCCAGCTGTCCTTCATGACCAAACGACATGTGCAGCGGGCCCTTCGCGACCTCACCATCGGG |
| droWil1 | TGCTGATGGCCATCATGTCGGGCCTCAATCTACTCTGTCGCCTGGCCATGATGCTGAGCAAGAGTGTGCGTGAGCAAATGATTCGCAGTCAATTGCGTTTTATGCCCAAACGGCATATCCAACGAGCTCTCAGGGATCTAACCATTGGC |
| droVir3 | TGCTGATGGCCTTCATGTCCGGCCTGAACCTGCTCTGCCGTCTGGCCATGATGCTCAGCCGGACGCTGCGTGAGCTGATGATACGCAGCCAATTGCGCTTCATGACCAAGCAGCATGTACAGCGCGTTTTCCGGGATCTAACCATCGGT |
| droMoj3 | TGCTGATGGCCTTCATGTCCGGCCTGAATCTGCTCTGTCGCCTGGCCATGATGCTCAGCCGGACGCTGCGCGAGCTGATGATACGCAGCCAATTGCGCTATATGACCAAACAGCATGTGCAGCACGTCTTCAGGGATCTGACCATCGGC |
| droGri2 | TCCTCATGGCGATCATCGCTGGCCTAAATCTGCTCTGTCGCCTAACCATGATGGTCAGCCGGACGCTGCGCGAACTGATGATTCGCAGCCAGTTGCGATTCATGACCAAAGAGCATGTGCAGCGCATCTTTAGCCATTTAACCATCGGC |
| dm3 | droSim1 | droSec1 | droYak2 | droEre2 | droEug1 | droBia1 | droTak1 | droEle1 | droRho1 | droFic1 | droKik1 | droAna3 | droBip1 | dp4 | droPer1 | droWil1 | droVir3 | droMoj3 | droGri2 | |
| RNAfold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| UNAFold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Species | Alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 04/20/2012 at 05:29:02 EDT