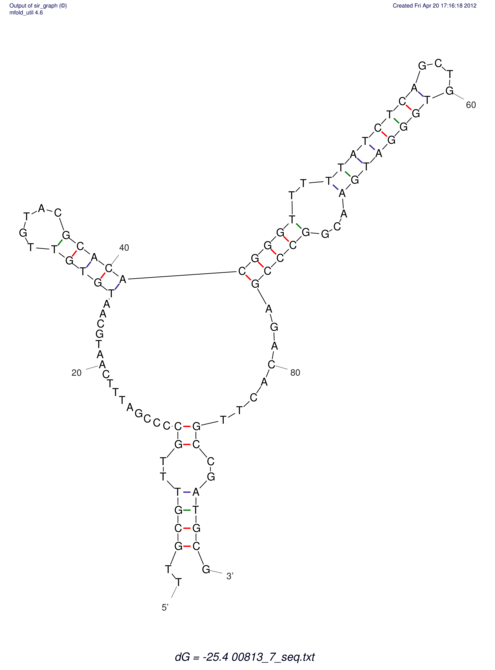
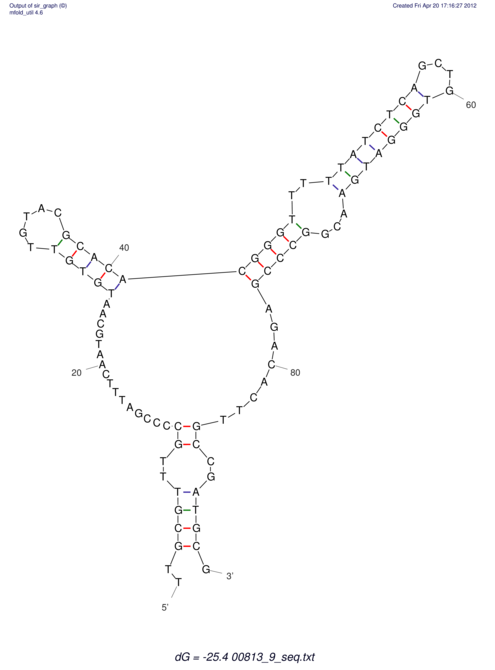
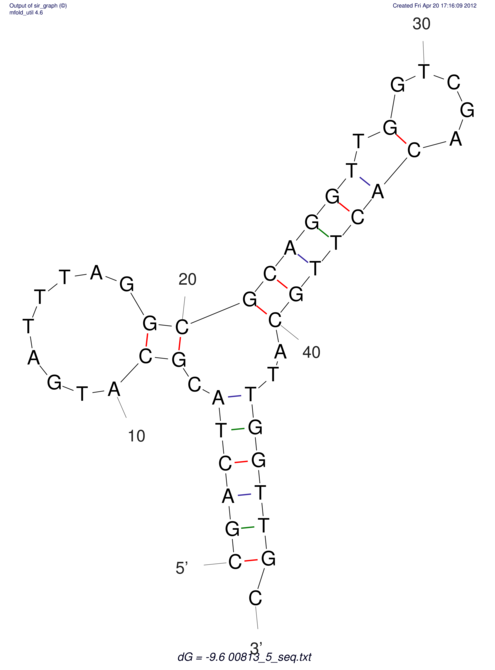
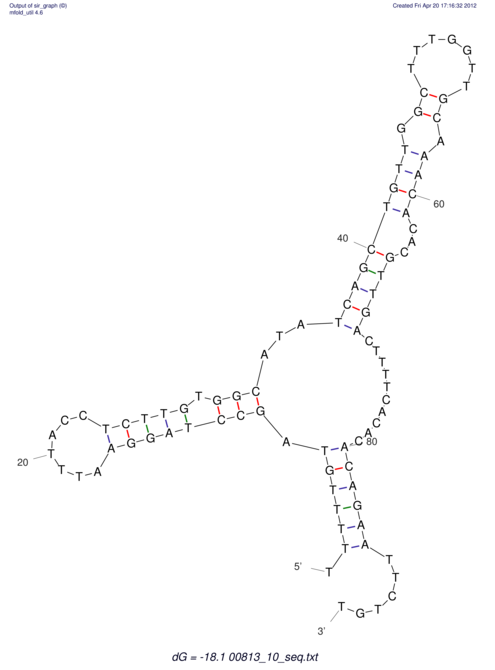
| dm3 |
chrX:15213971-15214085 - |
|
1 |
CTTTTTT-TTCT----CTG----CTCG--------------------------TTGTATTTTACGCACTCTGCTGGCAGCGCTGTCG------GCGTCGCTGCCAGCTGCCAGCAGC------------------------------------GAGCG--------------GT--ACACTTGCAACTA---------TCCTGTGCACCCT---ACAATT |
| droSim1 |
chrX:11736931-11737042 - |
LASTZ |
4 |
CTTTTTT-TTTTTTCTCTG----CTCG--------------------------TTGCATTTTACGCACTCTGCTGGCAGCGACGCCG------ACAGCGCTGCCAGCA-------GC------------------------------------GAGCG--------------GT--ACACTTGCAACTA---------TCCTGCGCACCCT---ACAATT |
| droSec1 |
super_56:101687-101796 - |
LASTZ |
4 |
CTTTTTT-TTTT--CTCTG----CTCG--------------------------TTGCATTTTACGCACTCTGCTGGCAGCGACGCCG------ACAGCGGTGCCAGCA-------GC------------------------------------GAGCG--------------GT--ACACTTGCAACTA---------TCCTGCACACCCT---ACAATT |
| droYak2 |
chrX:9414156-9414285 - |
LASTZ |
5 |
TTTTTTT-------CTCTG----CTCGTTTT---TCTCTCTTTTTCTTCTCGCCTGCATTTTACGCACTCTGCTAGCAGCGACGCCG------ACTGCGCTGCCAACA-------GC------------------------------------AAGCG--------------GT--ACACTTGCAACTA---------TTCTATGCACCCT---ACAATT |
| droEre2 |
scaffold_4690:11807313-11807440 + |
LASTZ |
5 |
TTTTTTC-T-------CCG----CTCGTTTT---TCTCTCCTTTTCTGCTCGCCTGCATTTTACGCACTCTGCTGGCAGCGACGCCG------ACTGCGCTGCAAACA-------GC------------------------------------GAGCG--------------GT--ACACTTGCAACTA---------TTCTGTGCACCCT---ACAATT |
| droEug1 |
scf7180000409533:350907-351023 - |
LASTZ |
5 |
TTTTTTC-TCT-----CTG----CTAGTTTTTTTT----------------GCTTGCATTTTACGCACTCTGCTGGCAGCGACGTCG------ACTGCGCTGCCAAGT-------GC------------------------------------AAGCG--------------GT--ACACTTGCAACTA---------TTCGTTGTACTCC----GAATT |
| droBia1 |
scf7180000299226:181007-181106 + |
LASTZ |
5 |
CTTCCCC-TCCC----CCG----CCCG------------------CCCCGCGGCAGCGTTTTACGCACTCTGCCGGCAGCGACGTCG------ACCGC-----------------------------------------------------------G--------------GT--ACACTTGCCACAA---------TTCCTTGTACCCC---CGAGAG |
| droTak1 |
scf7180000415381:822078-822183 - |
LASTZ |
5 |
TTTTTTT-------CTCAG----CC---------------------------CCTGCATTTTACGCACTCCGCTGGCAGCGACGTCG------ACTGCGCTGCCGACG-------TC------------------------------------AGGC-----------GGCGGT--ACACTTGCCACTA---------TTCTTTGTACCC------CCGT |
| droEle1 |
scf7180000491023:1386855-1386967 + |
LASTZ |
5 |
TTTTTTT-TTTT----TTGGGG-----TTTTTTTT----------------GCCTGCATTTTACGCACTCTGCTGGCAGCGACGTCG------ACTGCGACGCCGACT-------GC------------------------------------A-GCC--------------GT--ACACTTGCAACTA---------TTCGTTGTACCC------AATT |
| droRho1 |
scf7180000777097:282566-282669 + |
LASTZ |
5 |
TTTTTTT-TGCT---------TTTTC--------------------------GCTGCATTTTACGCACTCTGCTGGCAGCGACGTCG------ACTGCGACGGCGACT-------GC------------------------------------AGGCG--------------GT--ACACTTGCAACTA---------TTCTGTGTACCC------AATT |
| droFic1 |
scf7180000454038:448768-448871 - |
LASTZ |
5 |
TTTTTTG-TTGC----T----TTT----------T----------------GCCTGCATTTTACGCACACTGCTGGCAGCGACGTCG------ACTGCGACGCCGACC-------GC------------------------------------GAGT---------------GT--ACACTTTCAACTA---------TTTCATGTACCCC---ACA--- |
| droKik1 |
scf7180000302816:65604-65724 - |
LASTZ |
0 |
CTTTTGC-TTTG--CTATG----CCTGTGG-CTCTGC-----CCGCAATTTGTCCGTGCCGTACGCAC--TGCGGGCAGAGACGCCG------ACTGCGTAGCA------------------------------------------------------------------------CCACTTGCAACTGGGTTATTTCTT--TTGTATT-G---TCTTTT |
| droAna3 |
scaffold_13117:1427273-1427395 - |
LASTZ |
0 |
ATTTTTC-TTCC----C-A---TG----------CAACCTGTGTCCAT----TGTACGTTTTACGCACTCTCC--CCAGCAACACCG------CCGCCGATAGCGACT-------GC------------------------------------GACGAGTACGTTGGCAGCGG----CACTTGACTCT--------------TTTT--GCG---GCAATT |
| droBip1 |
scf7180000396425:686944-687063 + |
LASTZ |
0 |
GCATTCA-ACCT----GTG----TCCA--------------------T----TTTACGTTTTACGCACTCAGCC--CAGCAACAACGCCGTCGAGTGC------GACT-------GC------------------------------------GACGAGTACGCTGGCAGCGG----CACTTGACTCTT-------TTTGC--GGCAATTT---TCGACA |
| dp4 |
chrXL_group1e:12152468-12152588 - |
LASTZ |
0 |
CTTTAGC-TTTT-----TGCGT-----TTGCGTTTGCCCCGATTTCAATGCA-ATGTGTTGTACGCACACGGGTT---TTTATCTCA------GCTGTGGGATGAACG-------GC--------------------------------------CCG--------------AG--ACACTTGC--CGA---------TGCGGCGACCCC------AGGC |
| droPer1 |
super_14:1456573-1456693 + |
LASTZ |
0 |
CTTTAGC-TTTT-----TGCGT-----TTGCGTTTGCCCCGATTTCAATGCA-ATGTGTTGTACGCACACGGGTT---TTTATCTCA------GCTGTGGGATGAACG-------GC--------------------------------------CCG--------------AG--ACACTTGC--CGA---------TGCGGCGACCCC------AGGC |
| droWil1 |
Unknown |
|
0 |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
scaffold_12928:4095173-4095244 - |
LASTZ |
0 |
CTCTTTA-TTGC----A----ATG----------C----------------GAC-------TACGCATG-------------------------------------AT-------TT------------------------------------A-GGCGCAGGTTGGT----CG--ACACTTGCATTGG---------TTGCAAGT----------ACTT |
| droMoj3 |
scaffold_6680:3750941-3751063 - |
LASTZ |
0 |
TTTGTTT-TTGT----TTT----CTTT--T----------------TGTAGCCTAGGAATTTACC---TCTTGTGGCATATCAGCTG---TTGGCTTTGGTTGCAAAC-------AC------------------------------------ACGTT-------GACT---TTTCACACACAGAATTC---------TGT--TGAAATGTTTAACACTT |
| droGri2 |
scaffold_14853:566008-566117 + |
LASTZ |
0 |
CTCTTTTATTGC----A----ATG----------A----------GACTGCG-ACGCAT----------------------------------------------GAT-------TTGGGCGCAGGTCTATCACCACTTGCACTGACGACTGCTTTTG--------------CT--TTTGCTACTACTT---------TTACTTGCCAGCG---GAACTT |