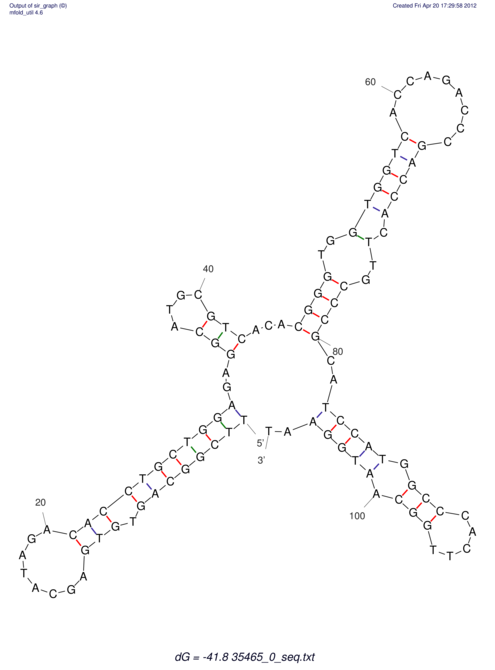
| dm3 |
chr3R:26876804-26876940 - |
NA |
GCACTCCCAGTCCCGGTCGTAGATGTGGGCGTAGGTGGCGGCGGGCGTGGCGTGCGTGATCCTGGTTGTGGTGACCACGCCGGTGCGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTCTGGACCCTGCCTTGCT |
| droSim1 |
chr3R:26523820-26523956 - |
13 |
GCACTCCCAGTCGCGGTCGTAGATGTGGGCGTAGGTGGCGGCGGGCGTGGCGTGGGTGATCCTGGTTGTGGTGACCACGCCCGTGCGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTCTGGACCCTGCCTTGTT |
| droSec1 |
super_4:5701489-5701625 - |
11 |
GCACTCCCAGTCGCGGTCGTAGATGTGGGCGTAGGTGGCGGCGGGCGTGGCGTGTGTGATCCTGGTTGTGGTGACCACGCCGGTGCGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTCTGGACCCTGCCTTGCT |
| droYak2 |
chr3R:27815996-27816132 - |
12 |
GCACTCCCAATCCCGGTCGTAGATGTGGGCGTACGTGGCGGCAGGCGTGGCATGGGTGATCCTGGTGGTGGTGACCACTCCAGTGCGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTTTGGACCCTACCTTGCT |
| droEre2 |
scaffold_4820:1105506-1105642 + |
5 |
GCACTCCCAATCCCGGTCGTAGATGTGGGCGTACGTGGCGGCAGGCGTGGCATGGGTGATCCTGGTTGTGGTGACCACTCCAGTACGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTTTGGACCCTGCCCTGCT |
| droEug1 |
scf7180000409166:158947-159083 + |
2 |
GCACTCCCAATCTCTGTCGTAGATGTGGGCGTAGGTGGCGGCAGGCGTTGCATGGGTGATCCTGGTGGTGGTGACAATACCAGTGCGCTTGCCCTCCTTTTGGGCCCACTCCATCACGCTTTGAACCCTGCCCTGTT |
| droBia1 |
scf7180000302113:442836-442972 + |
2 |
GCACTCCCAATCCCTGTCGTAAATATGGGCGTAGGTGGCCGCAGGCGTGGCGTGGGTGATCCTGGTGGTGGTGACGATTCCCGTGCGCTTGCCCGCCTTCTGGGCCCACTCCATTACGTTCTGGATCCTGCCTTGCT |
| droTak1 |
scf7180000415236:217947-218083 - |
8 |
GCACTCCCAATCGCGGTCGTAGATGTGGGCGTAGGTGGCAGCGGGCGTGGCATGAGTGATCCTGGTGGTGGTGACAATTCCAGTGCGCTTGCCCTCCTTTTGTGCCCATTCCATGACGCTTTGGACCCTGCCCTGCT |
| droEle1 |
scf7180000491212:972484-972620 - |
11 |
GCATTCCCAATTCCTGTCGTAAATATGGGCGTAAGTGGCGGCGGGCGTGGCATGGGTGATCCTAGTGGTGGTGACTATTCCCGTGCGTTTGCCCAGCTTTTGGGCCCACTCCATGACGCTTTGAACGCGACCTTGCT |
| droRho1 |
scf7180000761687:129-265 + |
3 |
GCACTCCCAATCTCTGTCGTAAATGTGGGCGTAGGTGGCGGAGGGCGTGGCATGTGTGATCCTGGTGGTGGTGACTATTCCCGTGCGCTTGCCCTCTTTTTGGGCCCACTCCATGACGCTTTGAACCTGACCTTGCT |
| droFic1 |
scf7180000453782:423025-423161 + |
1 |
GCACTCCCAATCCCTGTCGTAGATGTGGGCGTAAGTGGCGGCGGGCGTGGCGTGGGTGATCCTGGTGGTGGTGACAATTCCGGTGCGCTTGCCCGCCTTCTGAGCCCACTCCATGATGCTTTCGACGCGACCTTGCT |
| droKik1 |
scf7180000302238:91277-91413 - |
5 |
GCACTCCCAGTCCCGGTTGTATATATGGGCATATGTGGCAGCGGGTGTGGCATGCGTGATCCTGGTCGTGGTGACTACTCCAGTGCGTTTTCCCTCCTTTTGGGCCCAATCCATAATGCTCTGGACGCGACCCGATT |
| droAna3 |
scaffold_12911:4046748-4046884 + |
1 |
GCACTCCCAATCCCGATCGTAGATGTGGGCGTAGGTTGCAGCTGGCGTGGCGTGGGTAATCCGAGTGGTGGTCACTATTCCAGTTCGCTTCTTCTCCTTTTGGGCCCACTCCATGACGCTCTGAACGCGTCCTTGCT |
| droBip1 |
scf7180000396640:894541-894677 + |
3 |
GCACTCCCAATCCCGATCGTAGATGTGGGCGTAGGTTGCAGCTGGCGTGGCATGGGTAATCCGAGTGGTGGTAACTATTCCAGTTCGCTTTTTCTCCTTTTGGGCCCACTCCATGACGCTCGCGACACGTCCTTGCT |
| dp4 |
chr2:271882-272018 + |
1 |
GCACTCCCAGTCGCGATCGTAGATGTGGGCGTAGGTGGCCGCAGGCGTGGCATGCGTAATCCTCGTTGTGGTCACGATTCCCGTGCGCTTGCCTTCCTTCTGGGCCCACTCCATGATAGTCTCAATCCGGCCCAGCT |
| droPer1 |
super_7:3056347-3056483 + |
7 |
GCACTCCCAGTCGCGATCGTAGATGTGGGCGTAGGTGGCCGCAGGCGTGGCATGCGTAATCCTCGTTGTGGTCACGATTCCCGTGCGCTTGCCTTCCTTCTGGGCCCACTCCATGATAGTCTCAATCCGGCCCAGCT |
| droWil1 |
scaffold_181108:531799-531935 - |
13 |
GCATTCCCAGTCCCGATCATATATATGCGCATAGGTTGAGGCAGGCGTGGCATGTGTAATTCTGGTAGTGGTAACAATTCCCGTTCGTTTACCCGCACGCTGGGCCCAATCCATAATGGATTCCACCCGACCCTGTT |
| droVir3 |
scaffold_12855:1470098-1470234 + |
1 |
GCACTCCCAGTCCCGATGATAGACACGCGCAAAGGTGGCTGCGGGCGTGGCGTGCGTAATGCGTGTGGTGGTCACCACGCCAGTGCGTTTGCCTTCACGCTGTGCCCAGTCCATGATGGAGTCGAGTCGACCCTGGC |
| droMoj3 |
scaffold_6540:27213622-27213758 + |
4 |
GCACTCCCAGTCCCGATGATAGATCCTTGCATAGGTGGCCGCAGGTGTGGCGTGTGTTATACGCGTGGTAGTCACGACGCCGGTCCGCTTGCCCTCACGCTGTGCCCAGTCCATGATGGAGTCGAGTCGACCCTGGC |
| droGri2 |
scaffold_14624:4037357-4037493 + |
2 |
GCATTCCCAGTCACGATTATAGATATTGGCATAGCTAGATGCCGGTGTGGCATGCGTAATCCTTGTGTTTGTCACCACGCCCGTCCGTTTGCCCACCCGCTGTGCCCAGTCCATGATGGAGTCGAGGCGACCCTGTG |