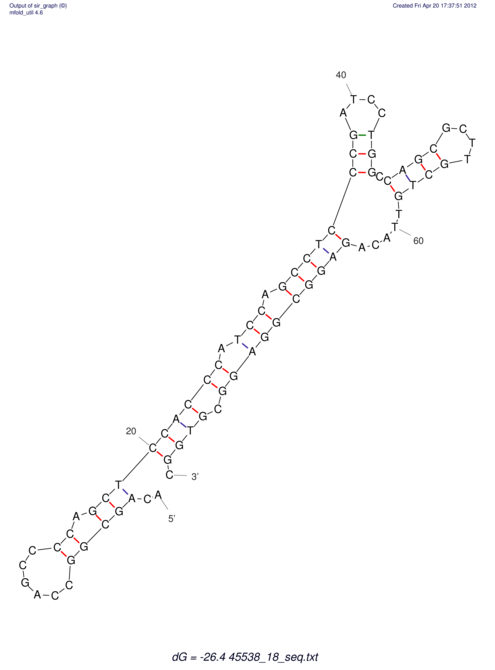
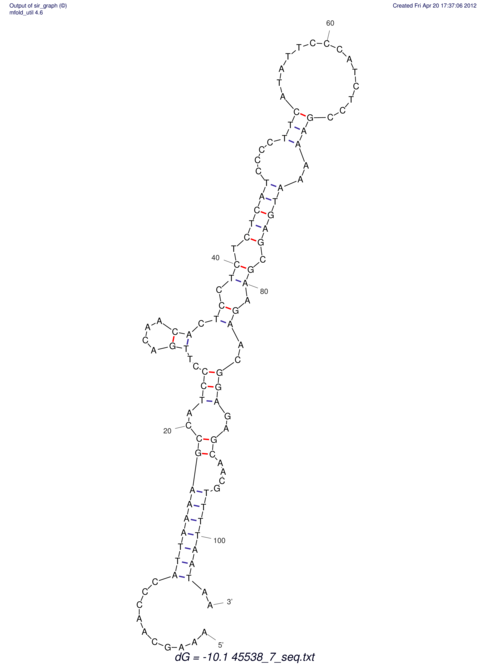
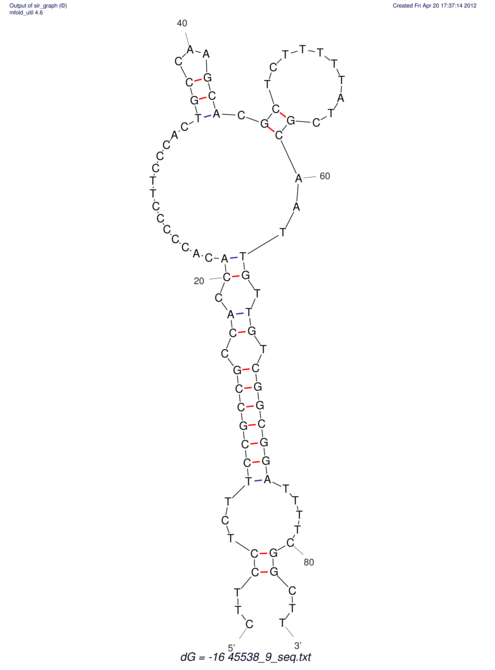
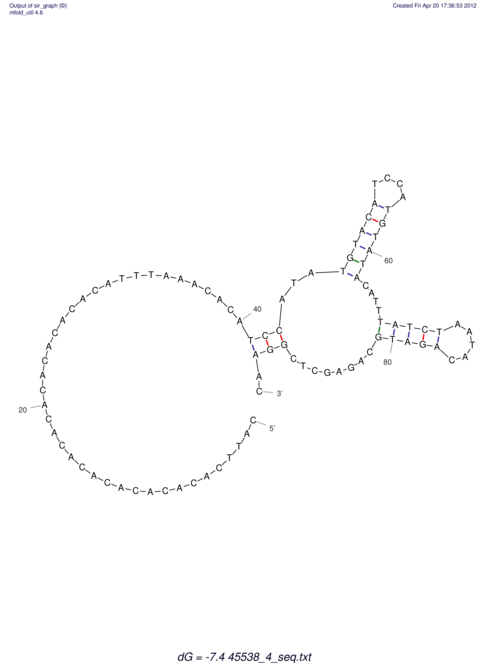
| CAAAATGTCCTTGATGACTGG-CATTCACACACA------C-A-C------ACACACA--CA-CACACATTTAAACACATCCATATGT---AC-------ATCCATGTATACATTTATCTAATA----------------------------------------------------CA-GATGCA-------G-----------AG------------CT---------------CGGAACTTCAAGT-------ATAAGGCA | Size | Hit Count | Total Norm | V117
Male body | V118
Embryo | V119
Head | M020
Head |
| ........................................C-A-C------ACACACA--CA-CATACAGT................................................................................................................................................................................................ | 20 | 19 | 0.11 | 0.05 | 0.05 | 0.00 | 0.00 |
| .......................TTCCACACACA------C-A-C------ACACACA--........................................................................................................................................................................................................... | 21 | 32 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| ............................CACACA------C-A-C------ACACACA--CA-CAC..................................................................................................................................................................................................... | 21 | 45 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ...............................ACA------C-A-C------ACAAACA--CA-CACACCT................................................................................................................................................................................................. | 22 | 8 | 0.13 | 0.00 | 0.13 | 0.00 | 0.00 |
| ...............................ACA------C-A-G------ACACACA--CA-CACACA.................................................................................................................................................................................................. | 21 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ................................CA------C-A-C------ACACACA--CA-CACAC................................................................................................................................................................................................... | 19 | 64 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 |
| .............................ACACA------C-A-C------ACACACG--CA-CACTC................................................................................................................................................................................................... | 22 | 51 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 |
| ..............................CACA------C-A-C------ACACACA--CA-CACAC................................................................................................................................................................................................... | 21 | 45 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ...............................ATA------C-A-T------ACACACA--CA-CACACAT................................................................................................................................................................................................. | 22 | 19 | 0.05 | 0.00 | 0.05 | 0.00 | 0.00 |
| .........................TCACACACG------C-G-C------ACACACA--C.......................................................................................................................................................................................................... | 20 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.25 |
| ................................CA------C-A-C------ACACACA--CC-CACACAGT................................................................................................................................................................................................ | 22 | 8 | 0.13 | 0.00 | 0.00 | 0.13 | 0.00 |
| ..............................CACA------C-A-C------ACACACA--CA-CACACA.................................................................................................................................................................................................. | 22 | 40 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| ..........................CACACACA------C-A-C------ACACACA--CA-CA...................................................................................................................................................................................................... | 22 | 40 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| .......................ATTCACACGCA------C-A-C------ACACGCA--........................................................................................................................................................................................................... | 21 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| ..............................CACA------C-A-C------ACTCACA--CA-TACACAT................................................................................................................................................................................................. | 23 | 7 | 0.14 | 0.14 | 0.00 | 0.00 | 0.00 |
| ..............................CACA------C-A-C------TCACACC--CA-CACAC................................................................................................................................................................................................... | 21 | 51 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 |
| ......................CACTCACACACA------C-A-C------ACACACA--TA-CACA.................................................................................................................................................................................................... | 28 | 19 | 0.05 | 0.00 | 0.05 | 0.00 | 0.00 |
| ............................................C------ACACACA--CA-CACATAGT................................................................................................................................................................................................ | 18 | 42 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 |
| ..........................................A-C------ACAGACG--CA-CACACAT................................................................................................................................................................................................. | 18 | 22 | 0.05 | 0.00 | 0.05 | 0.00 | 0.00 |
| ..........................CACACACT------C-A-C------ACCCACA--CA-C....................................................................................................................................................................................................... | 21 | 51 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 |
| ..........................CACACATA------C-A-C------ACACACA--TA-CACACA.................................................................................................................................................................................................. | 26 | 25 | 0.04 | 0.00 | 0.04 | 0.00 | 0.00 |
| .........................TTACACCCA------C-A-C------ACACACA--........................................................................................................................................................................................................... | 19 | 24 | 0.04 | 0.00 | 0.04 | 0.00 | 0.00 |
| .......................ATGCACGCACA------C-A-C------ACACACA--........................................................................................................................................................................................................... | 21 | 15 | 0.07 | 0.00 | 0.07 | 0.00 | 0.00 |
| ..............................CACA------C-A-C------TCACACC--CA-CACACA.................................................................................................................................................................................................. | 22 | 45 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ........................................C-A-C------ACACACA--CA-CACACATCTG.............................................................................................................................................................................................. | 22 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 |
| ...........................ACACACA------C-A-C------ACACGCA--CA-CTC..................................................................................................................................................................................................... | 22 | 51 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 |
| .............................ACACA------G-A-C------ACACACA--CA-CACA.................................................................................................................................................................................................... | 21 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ...........................ACACACA------C-A-C------ACACACA--CA-CACA.................................................................................................................................................................................................... | 23 | 36 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| .................................A------C-A-C------ACACACA--CA-CACATGT................................................................................................................................................................................................. | 20 | 37 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| ......................TATGCACACACA------C-A-C------ACACACA--........................................................................................................................................................................................................... | 22 | 26 | 0.04 | 0.00 | 0.00 | 0.04 | 0.00 |
| .................................A------C-A-C------ACACACA--CA-CACACATTGT.............................................................................................................................................................................................. | 23 | 6 | 0.17 | 0.17 | 0.00 | 0.00 | 0.00 |
| ..........................CACACACT------C-A-C------ACCCACA--CA-CA...................................................................................................................................................................................................... | 22 | 45 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| .........................TTCCACACA------C-A-C------ACACACA--CA-........................................................................................................................................................................................................ | 21 | 32 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| ..........................................A-C------ACACACA--CA-CACACATGT............................................................................................................................................................................................... | 20 | 37 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| ..........................CACACACA------C-A-C------ACACACA--CA-C....................................................................................................................................................................................................... | 21 | 45 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ...........................ACACAGA------C-A-C------ACACACA--CA-CA...................................................................................................................................................................................................... | 21 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ...................................................ACACACA--CA-CACACATTTGT............................................................................................................................................................................................. | 20 | 13 | 0.08 | 0.00 | 0.08 | 0.00 | 0.00 |
| ........................................C-A-C------ACACACA--CA-GACACAGT................................................................................................................................................................................................ | 20 | 15 | 0.07 | 0.00 | 0.07 | 0.00 | 0.00 |
| .........................TCACACACA------C-A-C------ATATACA--C.......................................................................................................................................................................................................... | 20 | 12 | 0.08 | 0.00 | 0.08 | 0.00 | 0.00 |
| ...........................AGACACA------C-A-C------ACACACA--CA-AACA.................................................................................................................................................................................................... | 23 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ...................................................ACACACA--CA-CACACATTGT.............................................................................................................................................................................................. | 19 | 14 | 0.07 | 0.00 | 0.07 | 0.00 | 0.00 |
| ..........................................A-C------ACACACA--TA-CACACATTTAAA............................................................................................................................................................................................ | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ............................CACACA------C-T-C------ACACCCA--CA-CAC..................................................................................................................................................................................................... | 21 | 51 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 |
| ............................CACACA------C-T-C------ACACCCA--CA-CACA.................................................................................................................................................................................................... | 22 | 45 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| .............................ACAAG------C-A-C------ACACACA--CA-CACACA.................................................................................................................................................................................................. | 23 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ................................CT------C-A-C------ACACACA--CA-AACACAT................................................................................................................................................................................................. | 21 | 10 | 0.10 | 0.00 | 0.10 | 0.00 | 0.00 |
| ...........................ACACACA------C-A-C------ACAAACA--CA-CACACAGT................................................................................................................................................................................................ | 27 | 5 | 0.20 | 0.20 | 0.00 | 0.00 | 0.00 |
| ........................................C-A-C------ACACACA--CA-CAGAGAT................................................................................................................................................................................................. | 19 | 43 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 |
| .............................ACACA------C-A-C------ACACACA--CA-CACACA.................................................................................................................................................................................................. | 23 | 36 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| ......................................................CACA--CA-CACACATCTATACACA........................................................................................................................................................................................ | 22 | 3 | 0.33 | 0.00 | 0.33 | 0.00 | 0.00 |
| ........................................C-A-C------ACACACA--CA-CACACATTGT.............................................................................................................................................................................................. | 22 | 7 | 0.14 | 0.14 | 0.00 | 0.00 | 0.00 |
| .............................AGACA------C-A-C------ACACACA--CA-CAAACA.................................................................................................................................................................................................. | 23 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ...........................ACAAGCA------C-A-C------ACACACA--CA-CACA.................................................................................................................................................................................................... | 23 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ............................................C------ACACACA--CA-CACACATTGT.............................................................................................................................................................................................. | 20 | 11 | 0.09 | 0.09 | 0.00 | 0.00 | 0.00 |
| .................................A------C-A-C------ACACACA--CA-CACACAGT................................................................................................................................................................................................ | 21 | 10 | 0.10 | 0.00 | 0.10 | 0.00 | 0.00 |
| ............................CACACA------C-A-C------ACACACA--CA-CACA.................................................................................................................................................................................................... | 22 | 40 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |