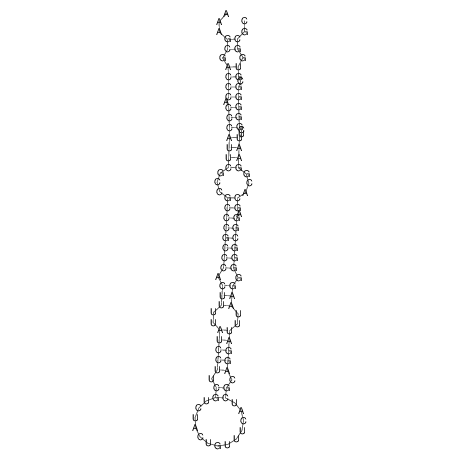
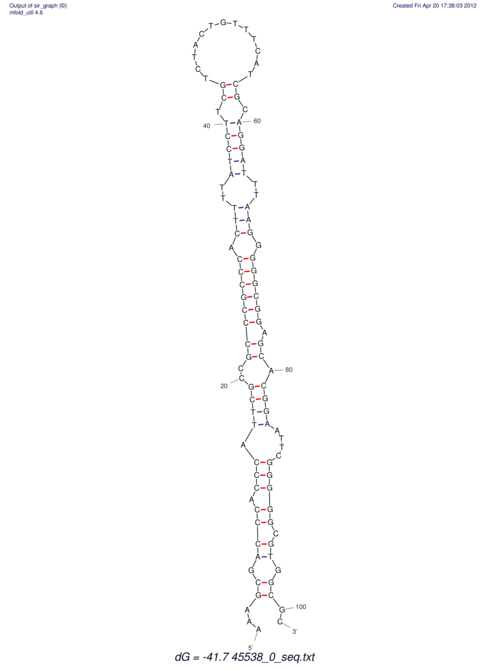
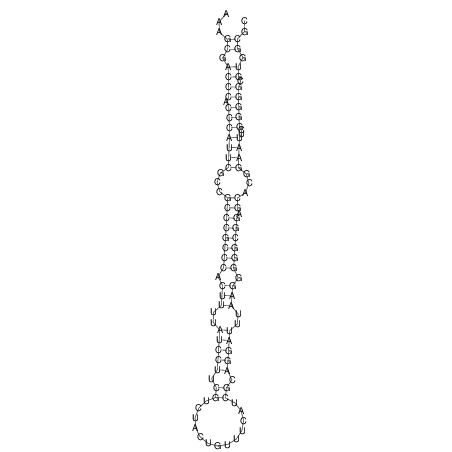
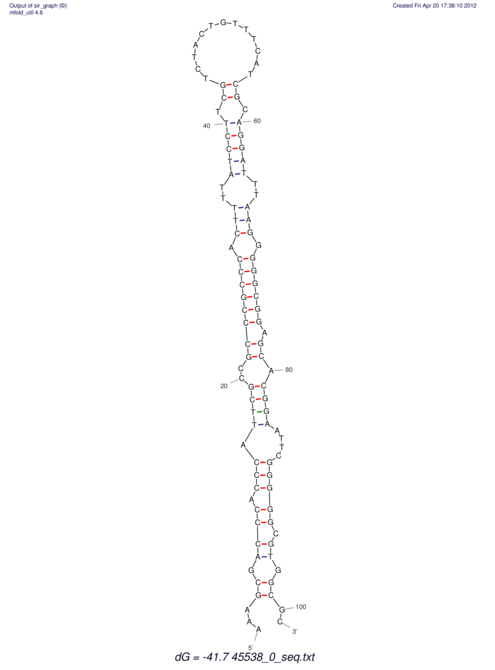
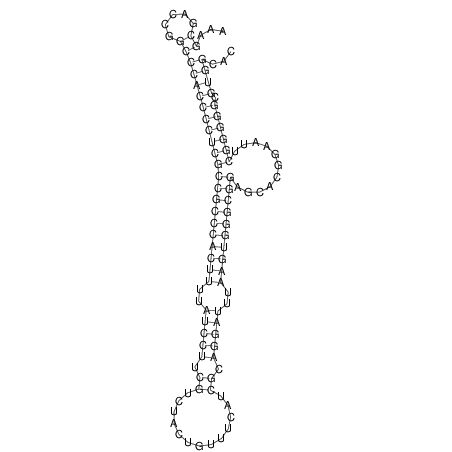
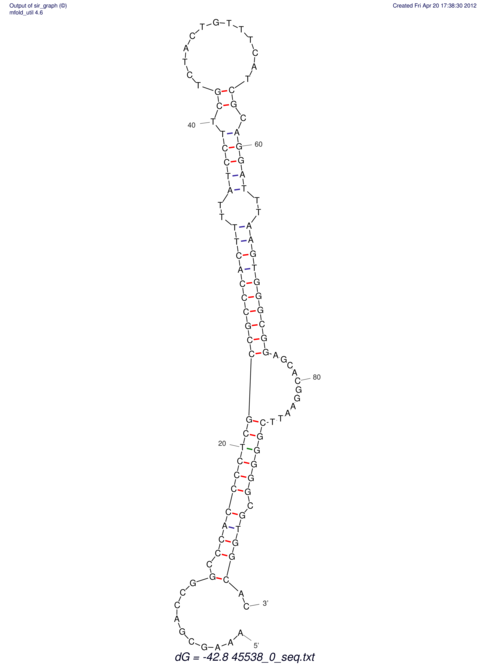
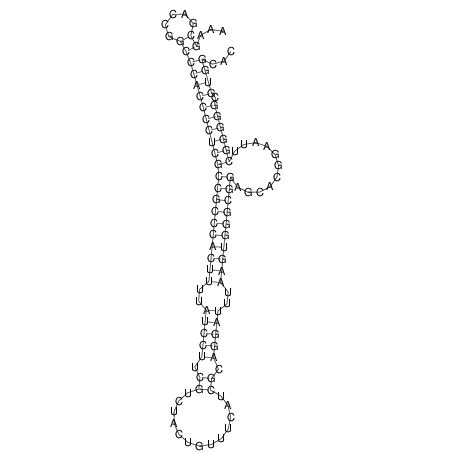
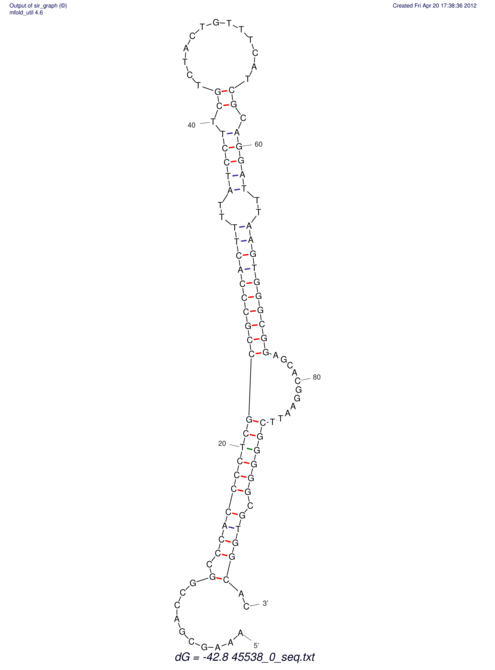
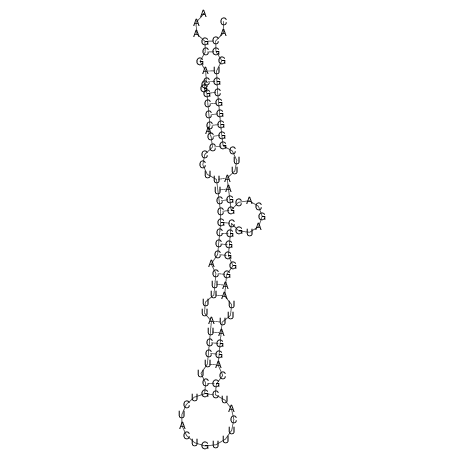
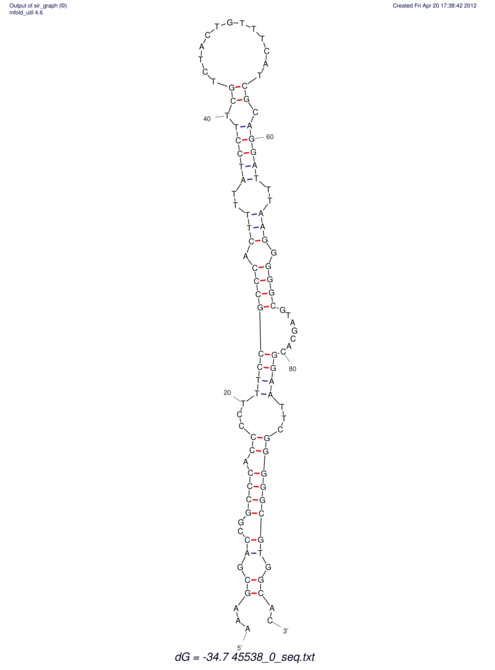
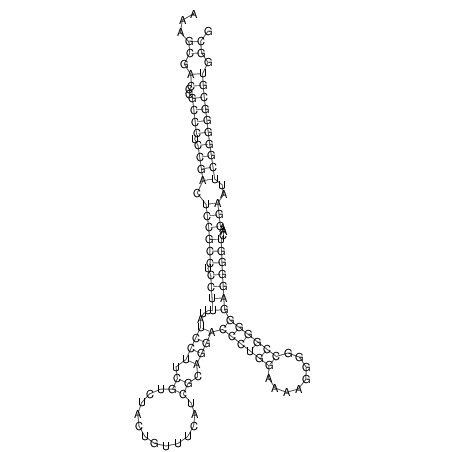
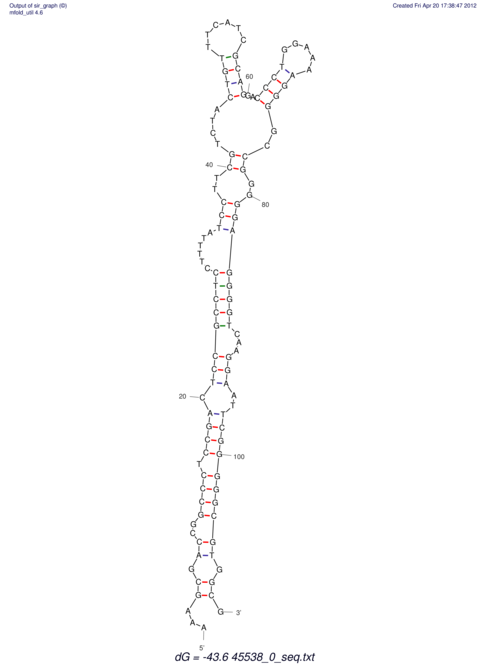
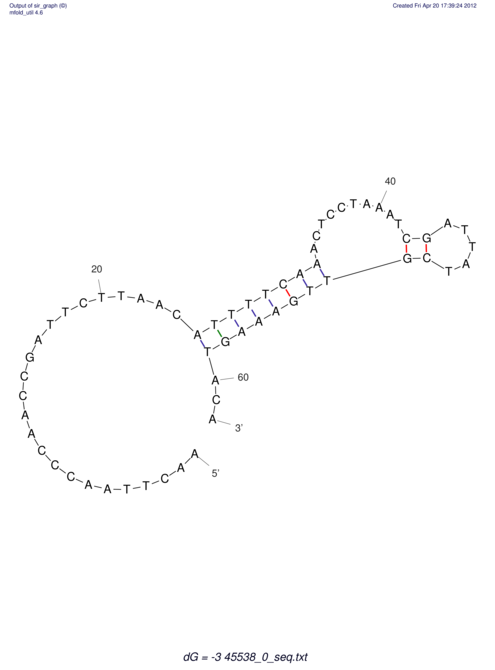
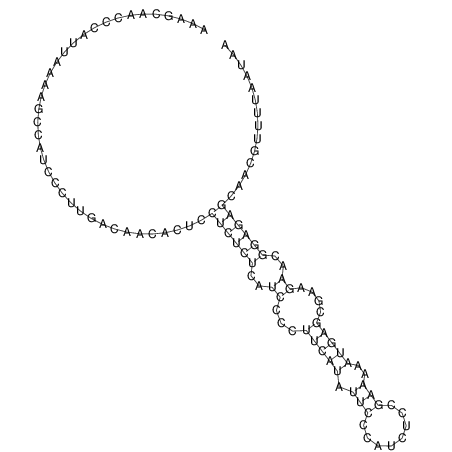
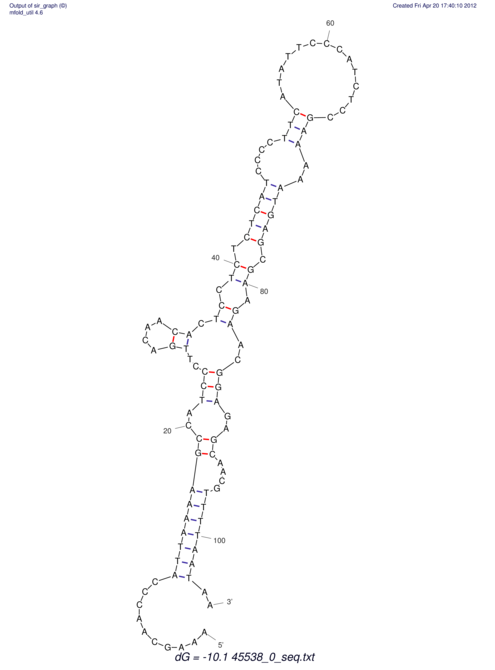
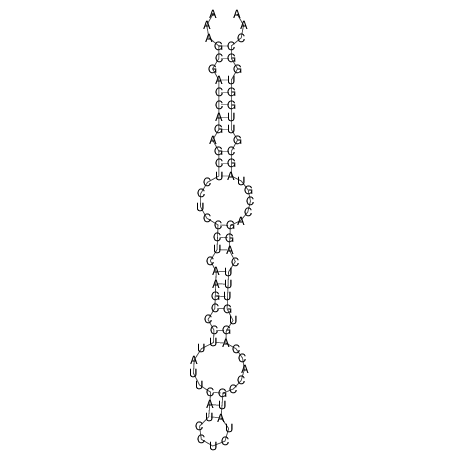
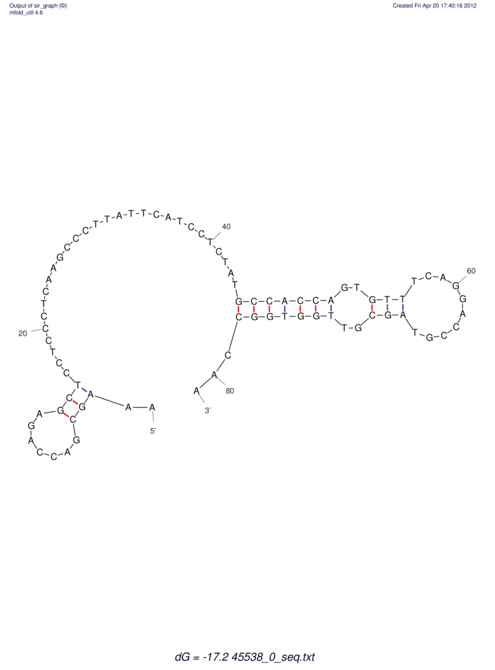
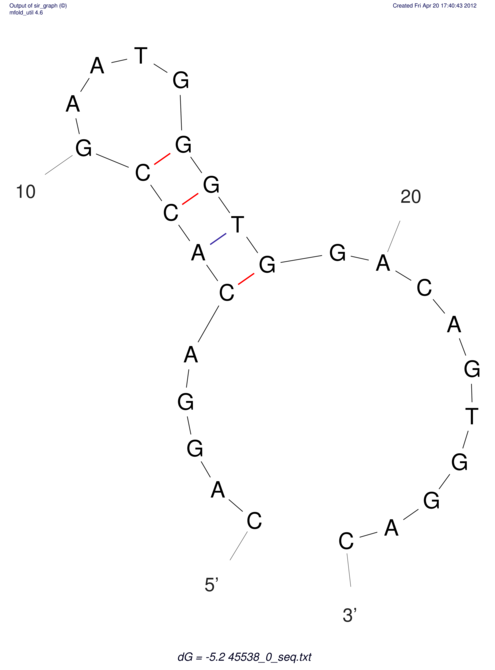
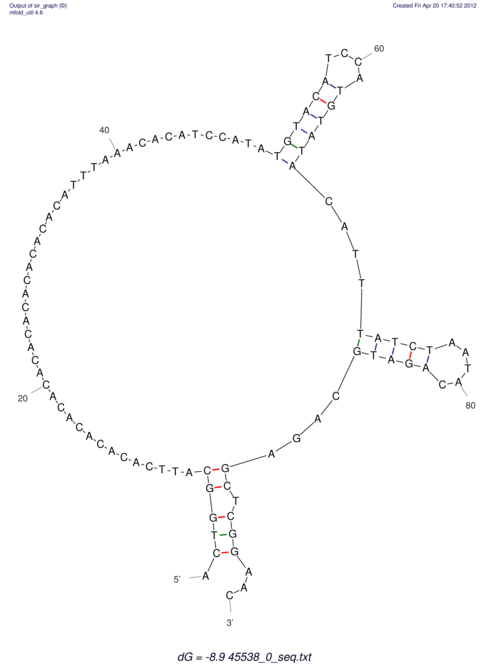
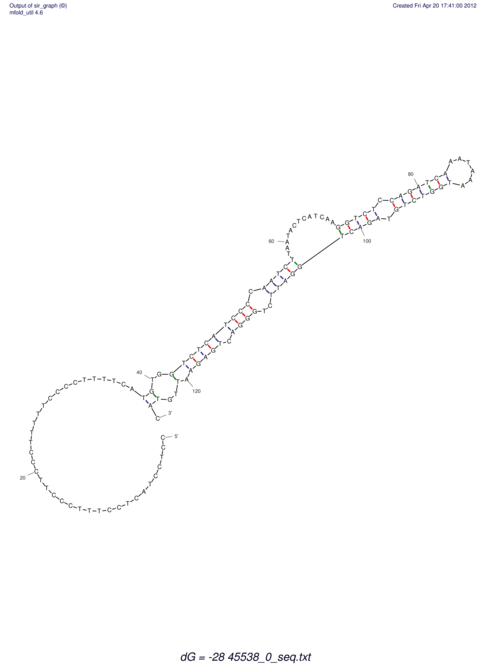
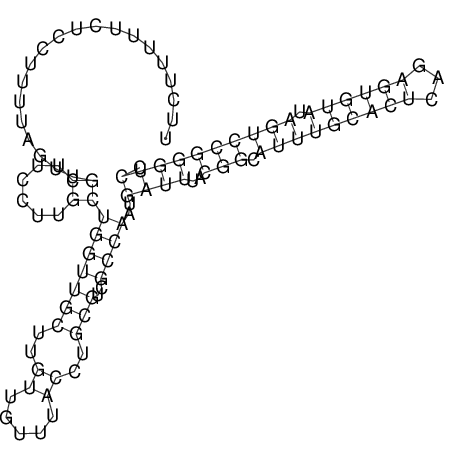
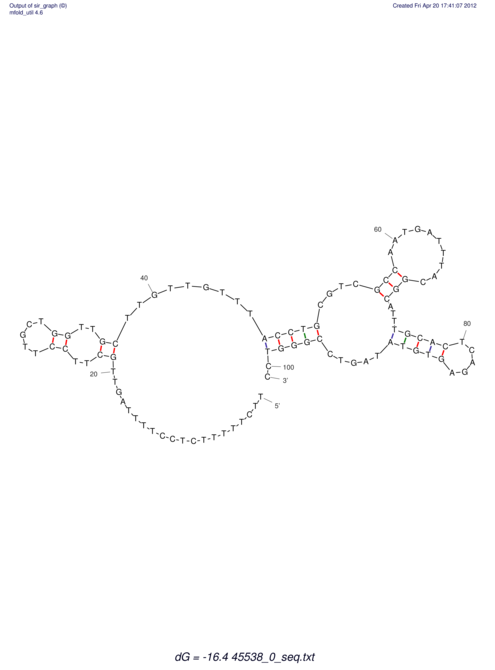
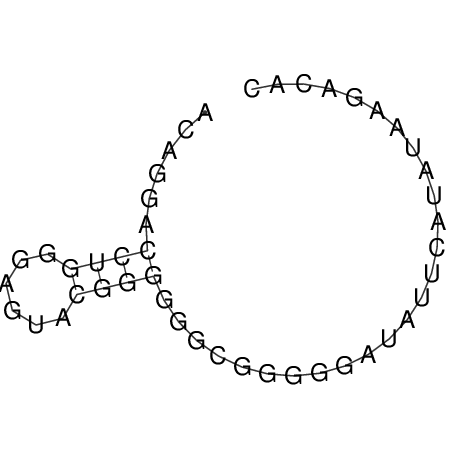
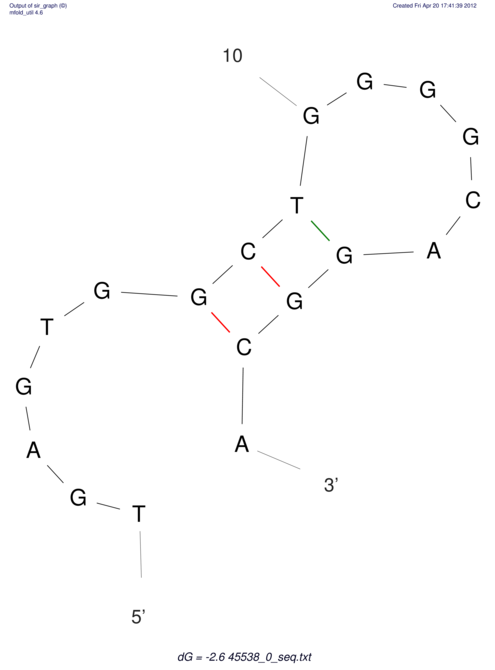
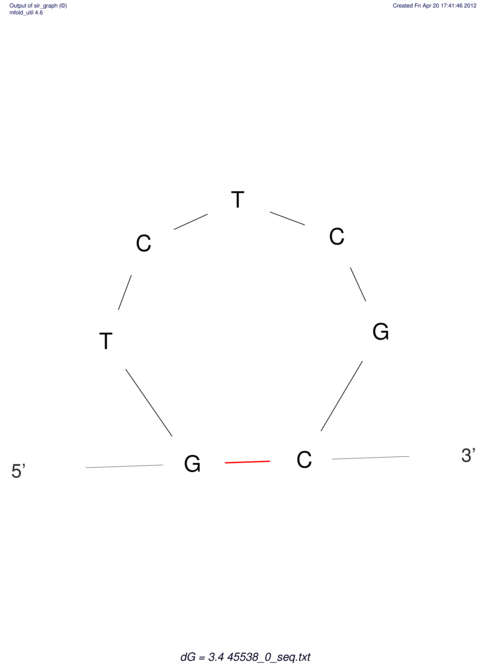
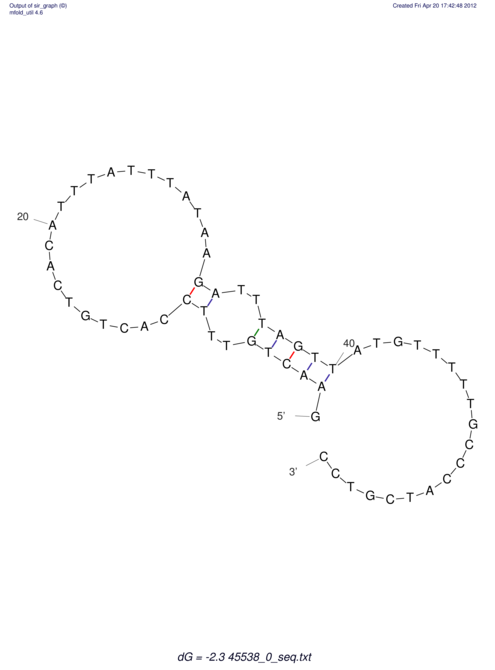
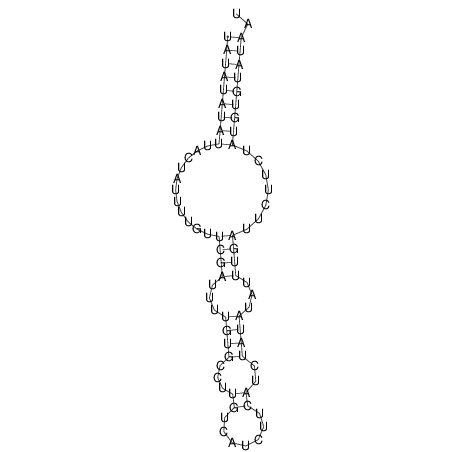
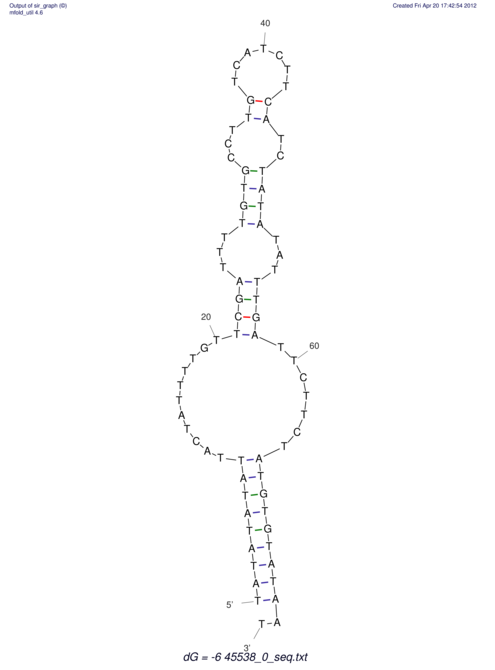
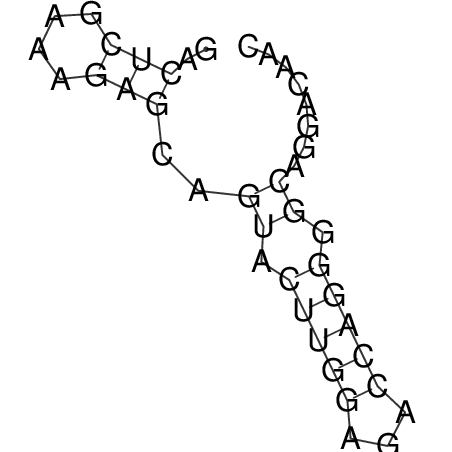
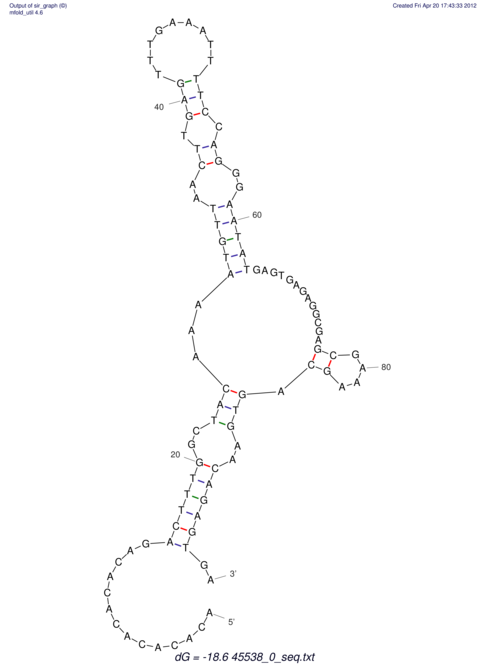
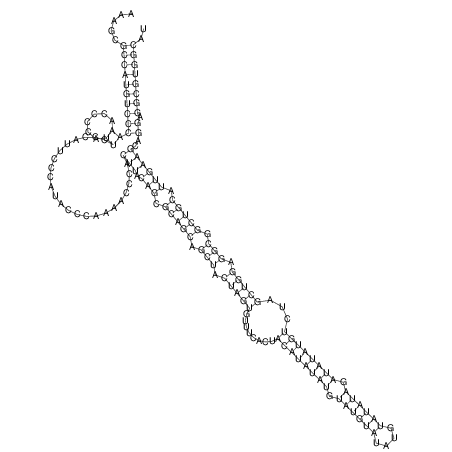
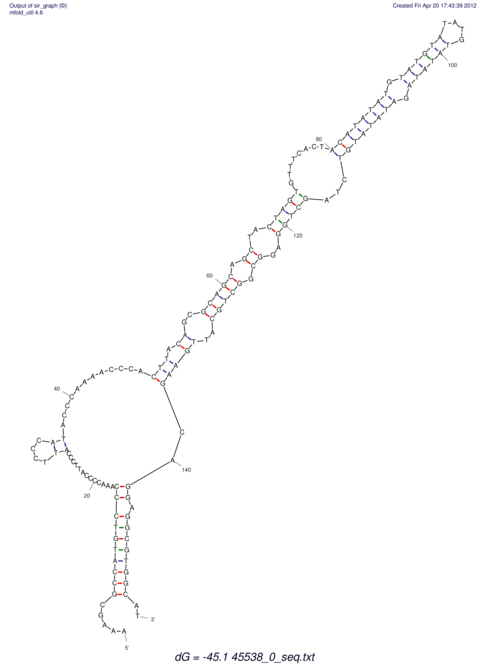
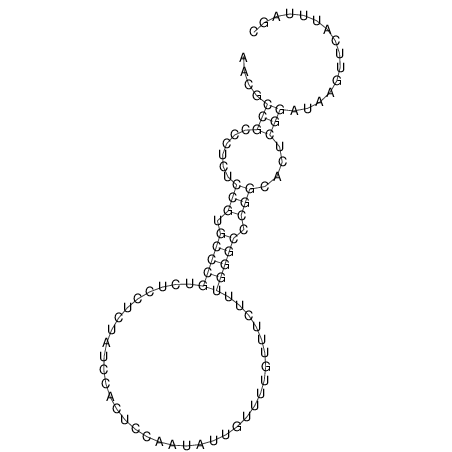
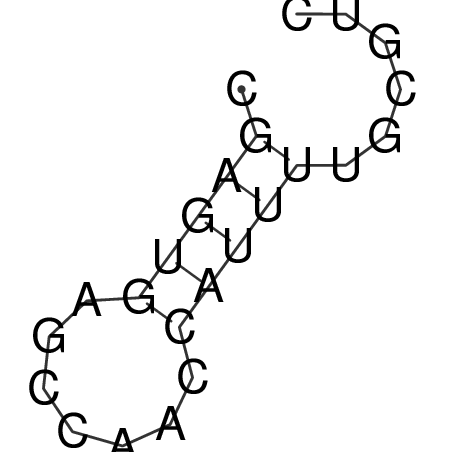
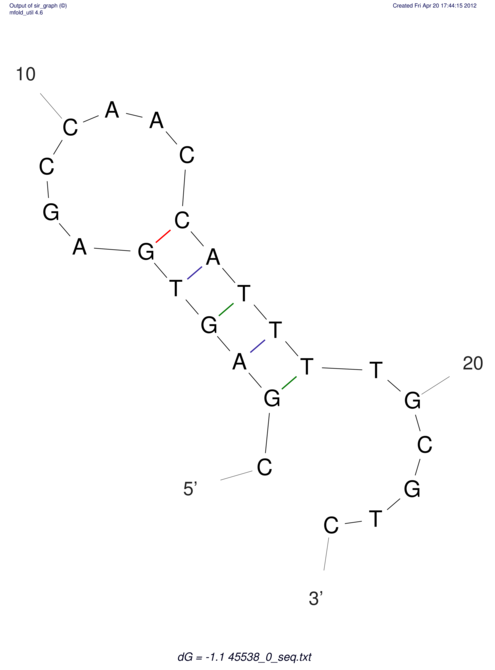
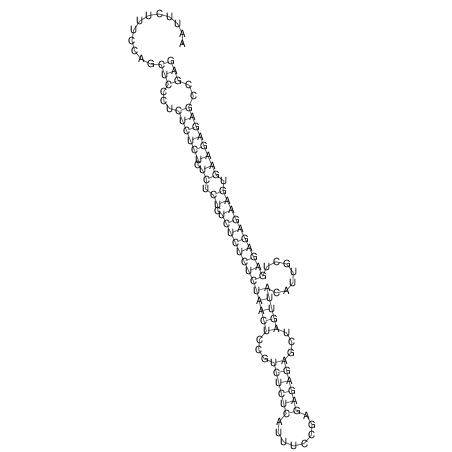
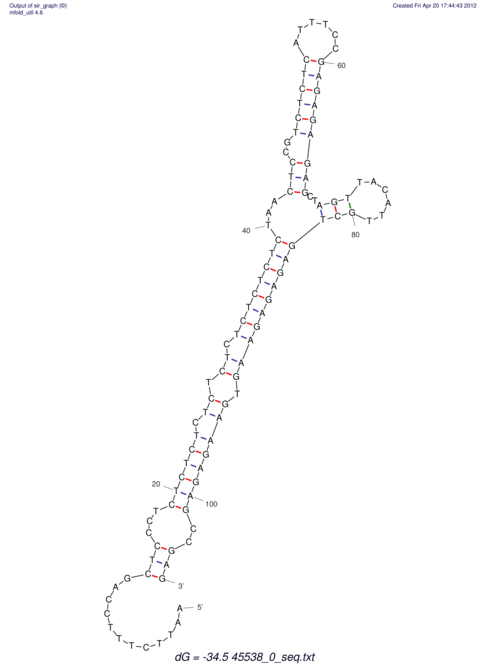
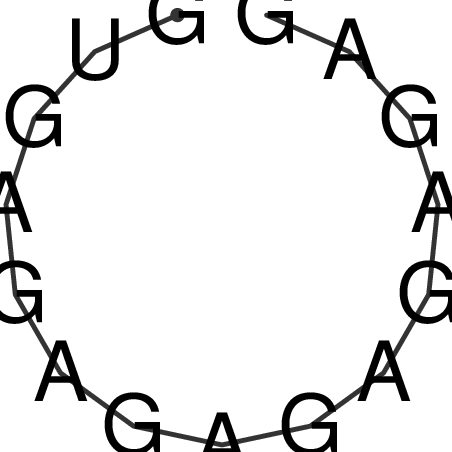
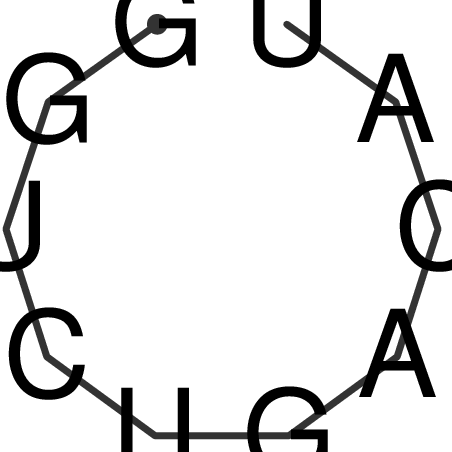
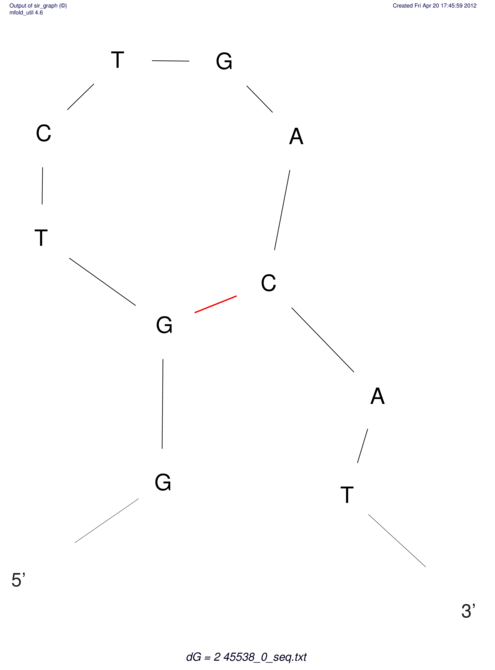
TAAAGTAATAAT-ATGAAA----GCGACCCACCCATTCCCCTTCCGCCCATATTTATCCTTCGTCTACTGTTTCATCGCAGGATTTAAGTGGGCGGAG------CACGGAATTCGGGGGCGTGGCGCACTGGGTAGCGGGTA
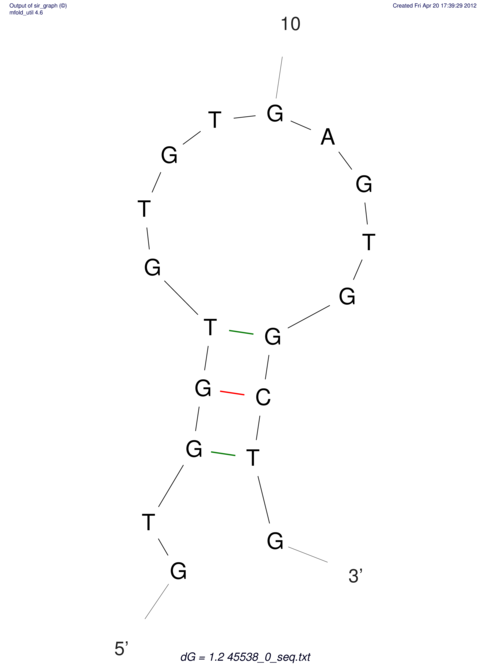
CAAAATGTCCTTGATGACTGGCATTCACACACACACA-CACACACACACACATTTAAACACATCCATATGTACATCC---------ATGTATACATTTATCTAATACAGATGC--AGAGCTCGGAACTTCAAGTATAAGGCA | Size | Hit Count | Total Norm | V117
Male body | V118
Embryo | V119
Head | M020
Head |
| .................................CACA-CACACACACATACAGT........................................................................................ | 20 | 19 | 0.11 | 0.05 | 0.05 | 0.00 | 0.00 |
| ......................TTCCACACACACACA-CACACA.................................................................................................. | 21 | 32 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| ...........................CACACACACA-CACACACACAC............................................................................................. | 21 | 45 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ..............................ACACACA-CAAACACACACACCT......................................................................................... | 22 | 8 | 0.13 | 0.00 | 0.13 | 0.00 | 0.00 |
| ..............................ACACAGA-CACACACACACACA.......................................................................................... | 21 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ...............................CACACA-CACACACACACAC........................................................................................... | 19 | 64 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 |
| ............................ACACACACA-CACACGCACACTC........................................................................................... | 22 | 51 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 |
| .............................CACACACA-CACACACACACAC........................................................................................... | 21 | 45 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ..............................ATACATA-CACACACACACACAT......................................................................................... | 22 | 19 | 0.05 | 0.00 | 0.05 | 0.00 | 0.00 |
| ........................TCACACACGCGCA-CACACAC................................................................................................. | 20 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.25 |
| ...............................CACACA-CACACACCCACACAGT........................................................................................ | 22 | 8 | 0.13 | 0.00 | 0.00 | 0.13 | 0.00 |
| .............................CACACACA-CACACACACACACA.......................................................................................... | 22 | 40 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| .........................CACACACACACA-CACACACACA.............................................................................................. | 22 | 40 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| ......................ATTCACACGCACACA-CACGCA.................................................................................................. | 21 | 1 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| .............................CACACACA-CTCACACATACACAT......................................................................................... | 23 | 7 | 0.14 | 0.14 | 0.00 | 0.00 | 0.00 |
| .............................CACACACT-CACACCCACACAC........................................................................................... | 21 | 51 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 |
| .....................CACTCACACACACACA-CACACATACACA............................................................................................ | 28 | 19 | 0.05 | 0.00 | 0.05 | 0.00 | 0.00 |
| ...................................CA-CACACACACACATAGT........................................................................................ | 18 | 42 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 |
| ..................................ACA-CAGACGCACACACAT......................................................................................... | 18 | 22 | 0.05 | 0.00 | 0.05 | 0.00 | 0.00 |
| .........................CACACACTCACA-CCCACACAC............................................................................................... | 21 | 51 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 |
| .........................CACACATACACA-CACACATACACACA.......................................................................................... | 26 | 25 | 0.04 | 0.00 | 0.04 | 0.00 | 0.00 |
| ........................TTACACCCACACA-CACACA.................................................................................................. | 19 | 24 | 0.04 | 0.00 | 0.04 | 0.00 | 0.00 |
| ......................ATGCACGCACACACA-CACACA.................................................................................................. | 21 | 15 | 0.07 | 0.00 | 0.07 | 0.00 | 0.00 |
| .............................CACACACT-CACACCCACACACA.......................................................................................... | 22 | 45 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| .................................CACA-CACACACACACACATCTG...................................................................................... | 22 | 7 | 0.14 | 0.00 | 0.00 | 0.00 | 0.14 |
| ..........................ACACACACACA-CACGCACACTC............................................................................................. | 22 | 51 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 |
| ............................ACACAGACA-CACACACACACA............................................................................................ | 21 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ..........................ACACACACACA-CACACACACACA............................................................................................ | 23 | 36 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| ................................ACACA-CACACACACACATGT......................................................................................... | 20 | 37 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| .....................TATGCACACACACACA-CACACA.................................................................................................. | 22 | 26 | 0.04 | 0.00 | 0.00 | 0.04 | 0.00 |
| ................................ACACA-CACACACACACACATTGT...................................................................................... | 23 | 6 | 0.17 | 0.17 | 0.00 | 0.00 | 0.00 |
| .........................CACACACTCACA-CCCACACACA.............................................................................................. | 22 | 45 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ........................TTCCACACACACA-CACACACA................................................................................................ | 21 | 32 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| ..................................ACA-CACACACACACACATGT....................................................................................... | 20 | 37 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| .........................CACACACACACA-CACACACAC............................................................................................... | 21 | 45 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ..........................ACACAGACACA-CACACACACA.............................................................................................. | 21 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ....................................A-CACACACACACACATTTGT..................................................................................... | 20 | 13 | 0.08 | 0.00 | 0.08 | 0.00 | 0.00 |
| .................................CACA-CACACACAGACACAGT........................................................................................ | 20 | 15 | 0.07 | 0.00 | 0.07 | 0.00 | 0.00 |
| ........................TCACACACACACA-TATACAC................................................................................................. | 20 | 12 | 0.08 | 0.00 | 0.08 | 0.00 | 0.00 |
| ..........................AGACACACACA-CACACACAAACA............................................................................................ | 23 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ....................................A-CACACACACACACATTGT...................................................................................... | 19 | 14 | 0.07 | 0.00 | 0.07 | 0.00 | 0.00 |
| ..................................ACA-CACACATACACACATTTAAA.................................................................................... | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ...........................CACACACTCA-CACCCACACAC............................................................................................. | 21 | 51 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 |
| ...........................CACACACTCA-CACCCACACACA............................................................................................ | 22 | 45 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ............................ACAAGCACA-CACACACACACACA.......................................................................................... | 23 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ...............................CTCACA-CACACACAAACACAT......................................................................................... | 21 | 10 | 0.10 | 0.00 | 0.10 | 0.00 | 0.00 |
| ..........................ACACACACACA-CAAACACACACACAGT........................................................................................ | 27 | 5 | 0.20 | 0.20 | 0.00 | 0.00 | 0.00 |
| .................................CACA-CACACACACAGAGAT......................................................................................... | 19 | 43 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 |
| ............................ACACACACA-CACACACACACACA.......................................................................................... | 23 | 36 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| ........................................CACACACACACATCTATACACA................................................................................ | 22 | 3 | 0.33 | 0.00 | 0.33 | 0.00 | 0.00 |
| .................................CACA-CACACACACACACATTGT...................................................................................... | 22 | 7 | 0.14 | 0.14 | 0.00 | 0.00 | 0.00 |
| ............................AGACACACA-CACACACACAAACA.......................................................................................... | 23 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ..........................ACAAGCACACA-CACACACACACA............................................................................................ | 23 | 51 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 |
| ...................................CA-CACACACACACACATTGT...................................................................................... | 20 | 11 | 0.09 | 0.09 | 0.00 | 0.00 | 0.00 |
| ................................ACACA-CACACACACACACAGT........................................................................................ | 21 | 10 | 0.10 | 0.00 | 0.10 | 0.00 | 0.00 |
| ...........................CACACACACA-CACACACACACA............................................................................................ | 22 | 40 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |