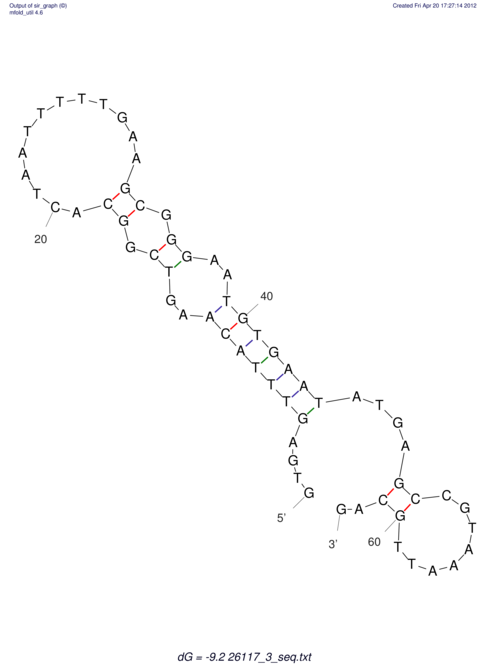
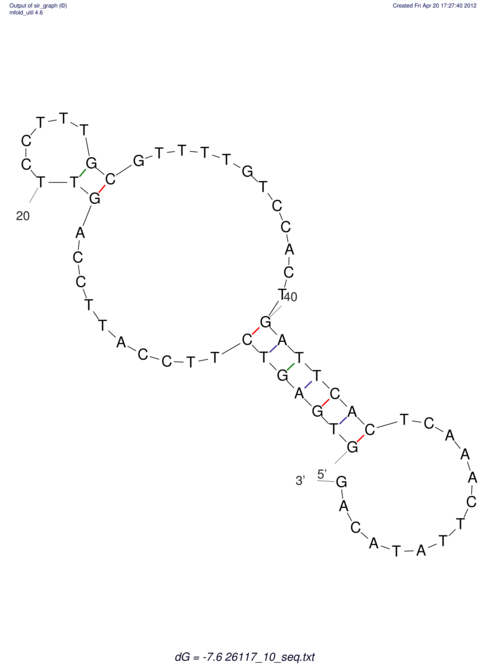
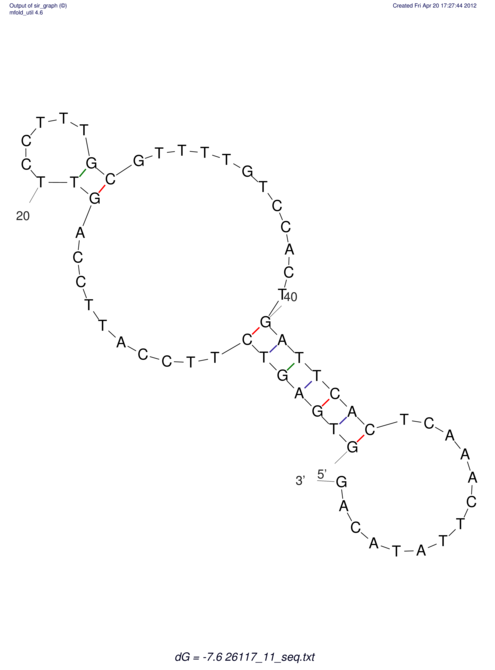
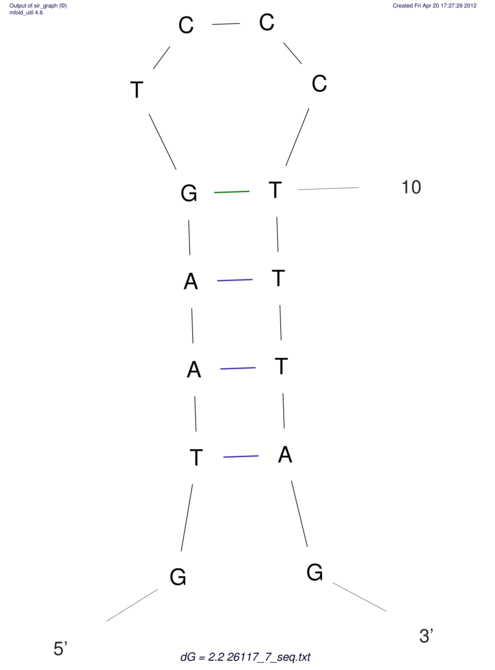
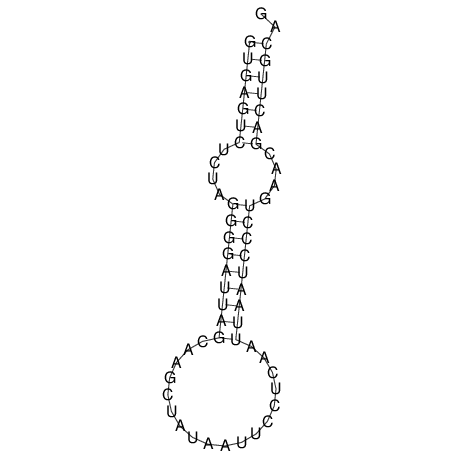
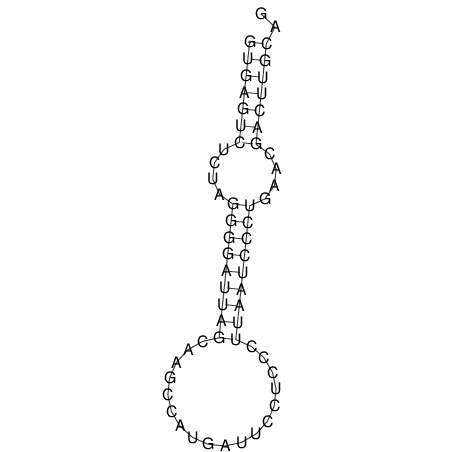
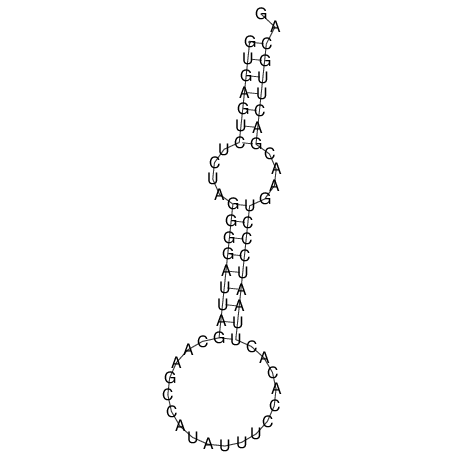
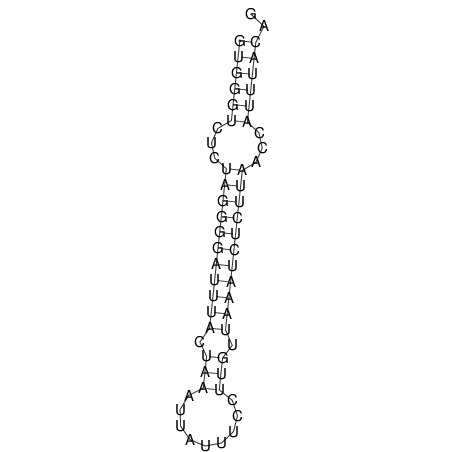
| TGGACATACTAAATGGTGAGTCTTT-----AGGGGATTA--GCAAGCCATAATTCC------------TC-ACTTAATCCCTGAA------------------CGACTTGCAGAGGCCGTTTTGGGTG | Size | Hit Count | Total Norm | V113
Male body | V114
Embryo | V115
Head |
| ..........................................................................TAATCCCTGAA------------------CGACTTGCAGT.............. | 22 | 1 | 226.00 | 134.00 | 70.00 | 22.00 |
| ...............GTGAGTCTTT-----AGGGGATTA--GC..................................................................................... | 21 | 1 | 39.00 | 19.00 | 18.00 | 2.00 |
| ..........................................................................TAATCCCTGAA------------------CGACTTGCAG............... | 21 | 1 | 38.00 | 24.00 | 8.00 | 6.00 |
| ..........................................................................TAATCCCTGAA------------------CGACTTGCAGA.............. | 22 | 1 | 17.00 | 14.00 | 3.00 | 0.00 |
| ...........................................................................AATCCCTGAA------------------CGACTTGCAGT.............. | 21 | 1 | 14.00 | 7.00 | 5.00 | 2.00 |
| ..........................................................................TAATCCCTGAA------------------CGACTTGCA................ | 20 | 1 | 7.00 | 4.00 | 0.00 | 3.00 |
| ..........................................................................TAATCCCTGAA------------------CGACTTGCAGTT............. | 23 | 1 | 4.00 | 3.00 | 0.00 | 1.00 |
| ...........................................................................AATCCCTGAA------------------CGACTTGCAG............... | 20 | 1 | 3.00 | 1.00 | 1.00 | 1.00 |
| ...............GTGAGTCTTT-----AGGGGATTA--G...................................................................................... | 20 | 1 | 3.00 | 2.00 | 0.00 | 1.00 |
| ..........................................................................TAATCCCTGAA------------------CGACTTGCGGT.............. | 22 | 1 | 2.00 | 1.00 | 1.00 | 0.00 |
| ..........................................................................TAATTCCTGAA------------------CGACTTGCAGT.............. | 22 | 1 | 2.00 | 1.00 | 1.00 | 0.00 |
| ...........................................................................AATCCCTGAA------------------CGACTTGCAGTT............. | 22 | 1 | 2.00 | 0.00 | 2.00 | 0.00 |
| ...............GTGAGTCTTT-----AGGGGATTA--GCA.................................................................................... | 22 | 1 | 2.00 | 0.00 | 2.00 | 0.00 |
| ...............GTGAGTCTTT-----AGGGGATTA--CC..................................................................................... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ...........................................................................AATCCCTGAA------------------CGACTTGCAGTA............. | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ..........................................................................TAATCCCTGAA------------------CGATTTGCAGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ...............GTGAGTCTTT-----AGGGGATTA--GCT.................................................................................... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ............................................................................ATCCCTGAA------------------CGACTTGCAGAGA............ | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ..........................................................................TAATCCCTGAA------------------CGACTTGTAGA.............. | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ...........................................................................TATCCCTGAA------------------CGACTTGCAGT.............. | 21 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ............................................................................ATCCCTGAA------------------CGACTTGCAGAT............. | 21 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ...................................................ATTCC------------TC-ACTTAATCCCTGAA------------------CGACTTG.................. | 28 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ..........................................................................CAATCCCTGAA------------------CGACTTGCAGT.............. | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ..........................................................................TAATCCCTGAA------------------CGGCTTGCAG............... | 21 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ...............GTGAGTCTTT-----AxGGGATTA--GC..................................................................................... | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| .........................................................................TTAATCCCTGAA------------------CGACTTGCAG............... | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ............................................................................ATCCCTGAA------------------CGACTTGCAGT.............. | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ..........................................................................TAATCCCTGAA------------------CGACTTGTAGT.............. | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ..........................................................................TAATCACTGAA------------------CGACTTGCAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| .........................................................................TTAATCCCTGAA------------------CGACTTGCAGT.............. | 23 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ..........................................................................TAATCTCTGAA------------------CGACTTGCAGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ...............GTGAGTCTTT-----AGGGGATT.......................................................................................... | 18 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ............................................................................ATCCCTGAA------------------CGACTTGCAGTT............. | 21 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ...............GTGAGTCTTT-----AGGGGATTC--G...................................................................................... | 20 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ...............GTGAGTCTTT-----AGGGGATTA--....................................................................................... | 19 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ..........................................................................TAATCCCTGAA------------------CGACTTGGAGT.............. | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| ..........................................................................TAATCCCTGAA------------------TGACTTGCAGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ..........................................................................TAATCCCTGAA------------------CGACTTGAAG............... | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 |
| .........................................................................TTAATCCCTGAA------------------CGACTTG.................. | 19 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ..........................................................................TAATCCCTGAA------------------CGACTTTCAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 1.00 |
| ...............GTGAGTCTTT-----AGGGGATTA--GCC.................................................................................... | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |
| ..........................................................................TAATCCCTGAA------------------CAACTTGCAGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 |