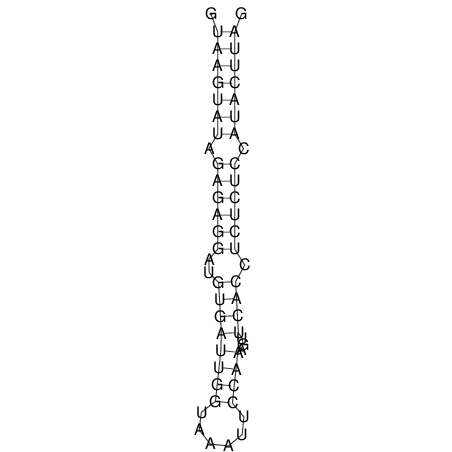
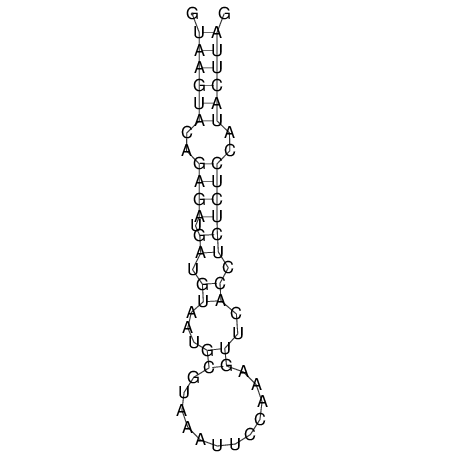
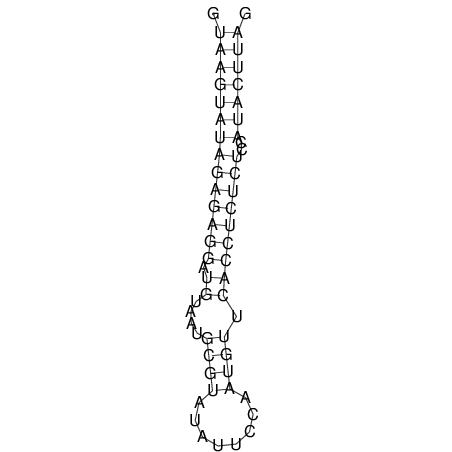
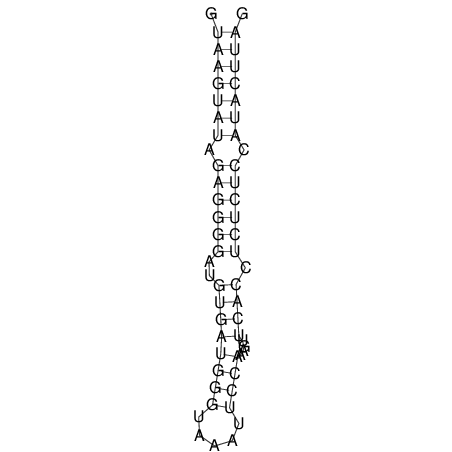
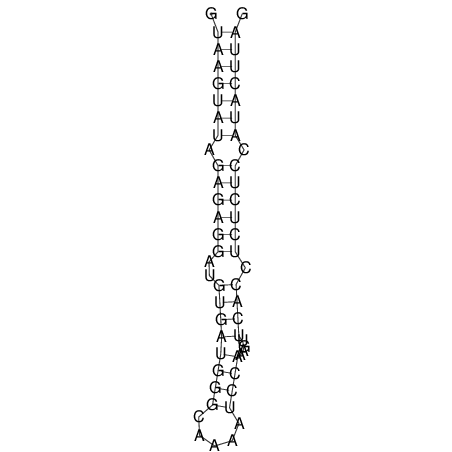
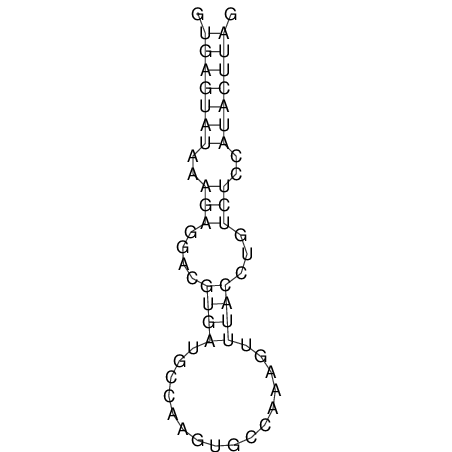
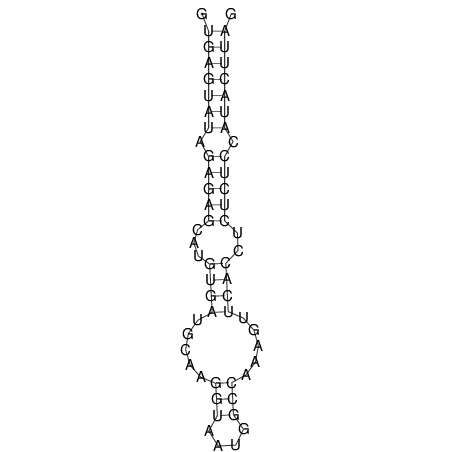
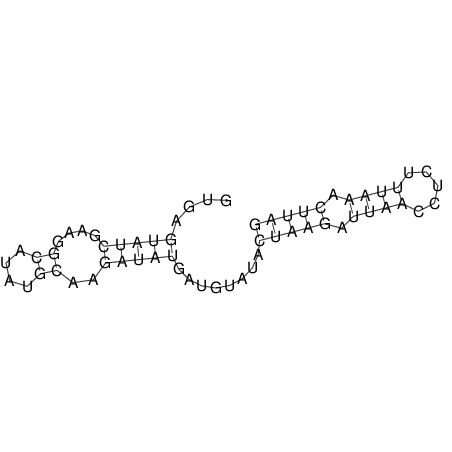
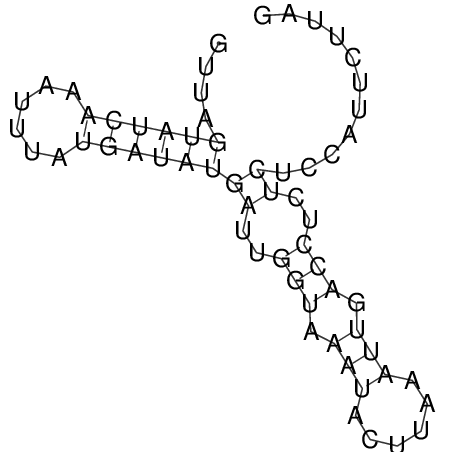
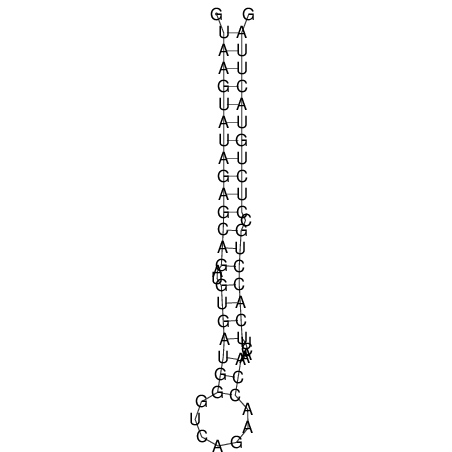
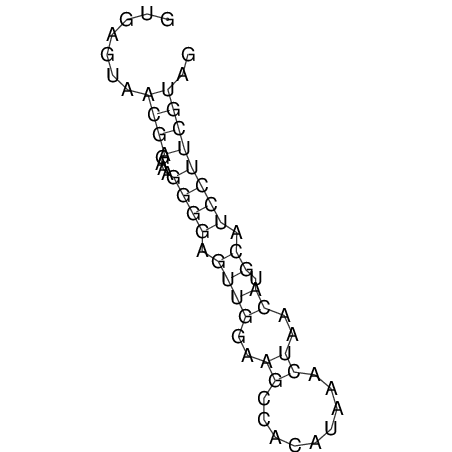
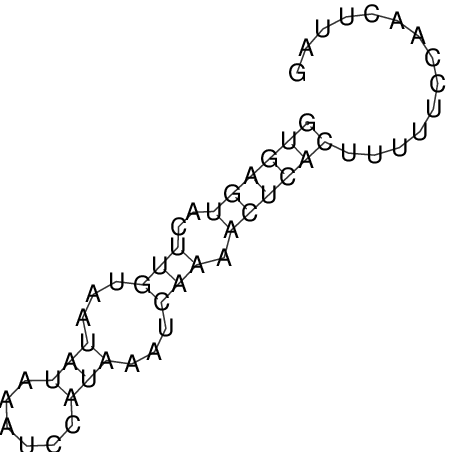
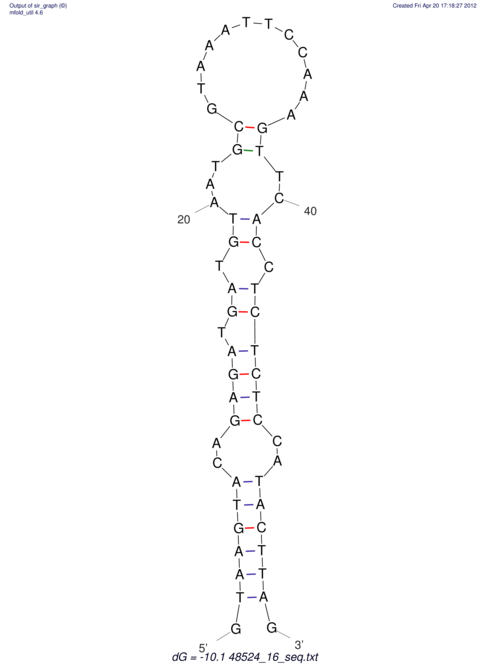
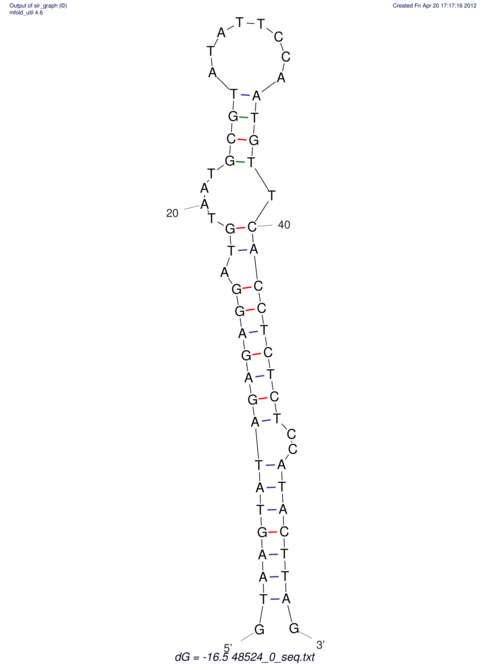
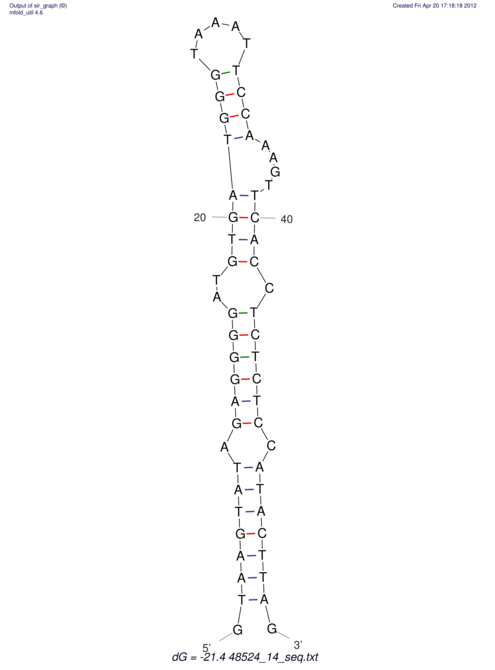
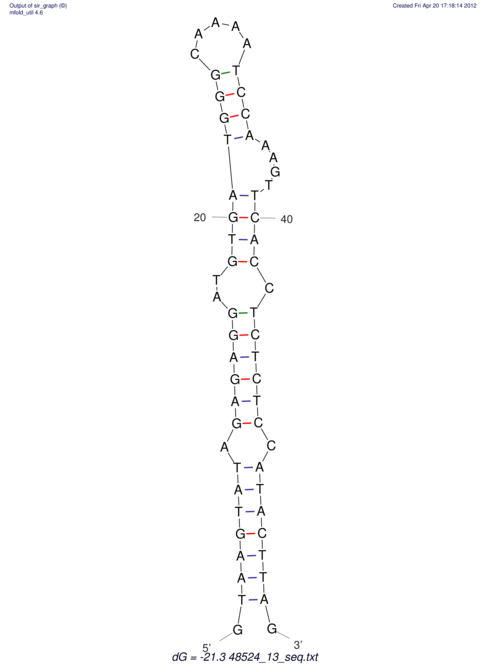
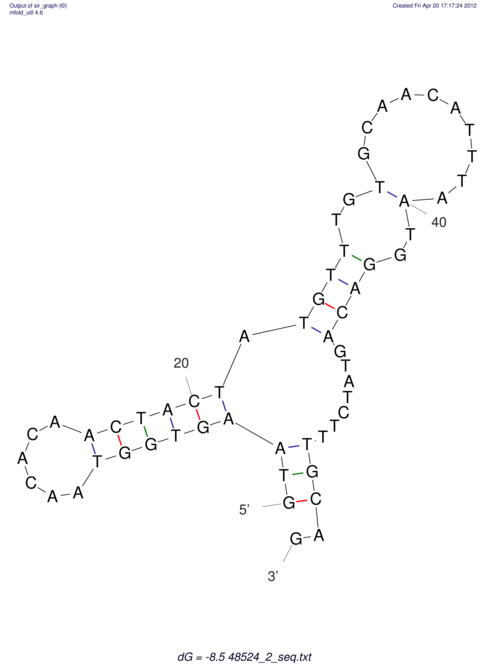
| CTCCAAGCTGTTTCAGTAAGTATAGAG---AG----GATGTGA-TG---GGCA-----------------AA-A---------------TCCAAAGTTCACCTCTCTCCA--TA-CTTAGTCGTCGAGGGGTTAT | Size | Hit Count | Total Norm | V040
Embryo | V060
Head | V047
Embryo | V108
Head |
| ................................................................................................TTCACCTCTCTCCA--TA-CTTAGT.............. | 22 | 1 | 866.00 | 21.00 | 58.00 | 0.00 | 787.00 |
| .................................................................................................TCACCTCTCTCCA--TA-CTTAGT.............. | 21 | 1 | 58.00 | 1.00 | 5.00 | 0.00 | 52.00 |
| .................................................................................................TCACCTCTCTCCA--TA-CTTAGTT............. | 22 | 1 | 36.00 | 3.00 | 4.00 | 0.00 | 29.00 |
| ................................................................................................TTCACCTCTCTCCA--TA-CTTAG............... | 21 | 1 | 22.00 | 0.00 | 1.00 | 0.00 | 21.00 |
| ...............GTAAGTATAGAG---AG----GATGTGA-TG---G..................................................................................... | 24 | 1 | 12.00 | 0.00 | 0.00 | 0.00 | 12.00 |
| ...............GTAAGTATAGAG---AG----GATGTGA-........................................................................................... | 21 | 1 | 11.00 | 0.00 | 1.00 | 0.00 | 10.00 |
| ................................................................................................TTCACCTCTCTCAA--TA-CTTAGT.............. | 22 | 1 | 9.00 | 0.00 | 0.00 | 0.00 | 9.00 |
| ...............GTAAGTATAGAG---AG----GATGTGA-TG---GGC................................................................................... | 26 | 1 | 8.00 | 0.00 | 0.00 | 0.00 | 8.00 |
| ...............GTAAGTATAGAG---AG----GATGTGA-TG---...................................................................................... | 23 | 1 | 7.00 | 0.00 | 1.00 | 0.00 | 6.00 |
| ................................................................................................TTCACATCTCTCCA--TA-CTTAGT.............. | 22 | 1 | 7.00 | 0.00 | 0.00 | 0.00 | 7.00 |
| ................................................................................................TTCACCTCTATCCA--TA-CTTAGT.............. | 22 | 1 | 6.00 | 0.00 | 0.00 | 0.00 | 6.00 |
| ................................................................................................TTCACCTATCTCCA--TA-CTTAGT.............. | 22 | 1 | 6.00 | 0.00 | 0.00 | 0.00 | 6.00 |
| ................................................................................................TTCACCTCTCTCCA--TA-CTTA................ | 20 | 1 | 5.00 | 0.00 | 1.00 | 0.00 | 4.00 |
| ................................................................................................TTAACCTCTCTCCA--TA-CTTAGT.............. | 22 | 1 | 4.00 | 0.00 | 0.00 | 0.00 | 4.00 |
| ...............GTAAGTATAGAG---AG----GATGTGA-TG---GG.................................................................................... | 25 | 1 | 4.00 | 1.00 | 0.00 | 0.00 | 3.00 |
| ................................................................................................TTCACCTCTCTCCA--TA-CTT................. | 19 | 1 | 4.00 | 0.00 | 0.00 | 0.00 | 4.00 |
| ................................................................................................TTCACCTCTCTACA--TA-CTTAGT.............. | 22 | 1 | 3.00 | 0.00 | 0.00 | 0.00 | 3.00 |
| ................................................................................................TTCACCTCTCTCCA--TA-ATTAGT.............. | 22 | 1 | 3.00 | 0.00 | 0.00 | 0.00 | 3.00 |
| ................................................................................................TTCACCTCTCTCCA--TA-CCTAGT.............. | 22 | 1 | 3.00 | 0.00 | 0.00 | 0.00 | 3.00 |
| ...................................................................................................ACCTCTCTCCA--TA-CTTAGT.............. | 19 | 1 | 3.00 | 1.00 | 1.00 | 0.00 | 1.00 |
| ................................................................................................TTCACCTCTCTCCA--TA-CTTAGA.............. | 22 | 1 | 3.00 | 0.00 | 1.00 | 0.00 | 2.00 |
| ................................................................................................TTCACCTCTCTCCA--TA-CTTATT.............. | 22 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| .................................................................................................TCACCTCTCTCCA--TA-CTTAGTA............. | 22 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| ................................................................................................TTCACCTCTTTCCA--TA-CTTAGT.............. | 22 | 1 | 2.00 | 0.00 | 1.00 | 0.00 | 1.00 |
| ................................................................................................TTCAACTCTCTCCA--TA-CTTAGT.............. | 22 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| ................................................................................................TTCACCTCTCTCCA--TA-CTTGGT.............. | 22 | 1 | 2.00 | 1.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCACCCCTCTCCA--TA-CTTAGT.............. | 22 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| ................................................................................................TTCACCTCTCTCCA--TA-CTTAGTT............. | 23 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| ................................................................................................TTCACCTCTCTCCA--TC-CTTAGT.............. | 22 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| ...............................................................................................CTTCACCTCTCTCCA--TA-CTTAGT.............. | 23 | 1 | 2.00 | 0.00 | 1.00 | 0.00 | 1.00 |
| ................................................................................................TTCGCCTCTCTCCA--TA-CTTAGT.............. | 22 | 1 | 2.00 | 0.00 | 1.00 | 0.00 | 1.00 |
| ................................................................................................TTCACCTCTCTCCA--TA-CTTAGC.............. | 22 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| ...............GTAAGTATAGAG---AG----GATGTGA-TC---GGC................................................................................... | 26 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............TTAAGTATAGAG---AG----GATGTG............................................................................................. | 20 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCACCTCTCTCTA--TA-CTTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCACCTCTCTCCC--TA-CTTAGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ................TAAGTATAGAG---AG----GATGTGA-TG---...................................................................................... | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .................................................................................................TCACCTCTCTCCA--TA-CTTAGTAA............ | 23 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTATAGAG---AG----GATGT.............................................................................................. | 19 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ............................................................................................AAAGTTCACCTCTCTCCA--TA-CT.................. | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .................................................................................................TCACCTCTCTCCC--TA-CTTAGT.............. | 21 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCACCTCTCTCCA--TC-CTTCGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ...............GTAAGTATAGAG---AT----GATGT.............................................................................................. | 19 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCACCTCTCTCCA--TA-CTTAGCA............. | 23 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTATAGAG---AG----GATGTG............................................................................................. | 20 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTATAGAG---AG----GATGTGA-T.......................................................................................... | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............................................................................................GTTCACCTCTCTCCA--TA-CTTAGT.............. | 23 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCAACTCTCTCAA--TA-CTT................. | 19 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCACCTCTCTACA--TA-ATTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCACACCTCTCCA--TA-CTTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................CTCACCTCTCTCCA--TA-CTTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .................AAGTATAGAG---AG----GATGTGA-TG---GGC................................................................................... | 24 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ..................................................................................................CACCTCTCTCCA--TA-CTTAGT.............. | 20 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .................................................................................................TCACCTCTCTCCA--TA-ATTAGT.............. | 21 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .................................................................................................TAACCTCTCTCCA--TA-CTTAGT.............. | 21 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .................................................................................................TCACCTCTCTCCA--TA-CTTAGTGT............ | 23 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCACCTCTCTCCA--TA-CTTAGG.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .................................................................................................TCACCTCTCTCCA--TA-CTTAG............... | 20 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTGACCTCTCTCCA--TA-CTTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTATAGAG---AG----GATTTGA-TG---GGC................................................................................... | 26 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............TTAAGTATAGAG---AG----GATGCGA-........................................................................................... | 21 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCAGCTCTCTCCA--TA-CTTAGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .................................................................................................TCACATCTCTCCA--TA-CTTAGTT............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTATAGAG---AG----GATGTGA-TC---...................................................................................... | 23 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTATAGAG---AG----GATGTCA-T.......................................................................................... | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .................................................................................................TCACCTATCTCCA--TA-CTTAGT.............. | 21 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................................................CCTCTCTCCA--TA-CTTAGT.............. | 18 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .................................................................................................TCACCTCTCTCCA--TA-CTTAGAT............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCACCTCTCTGCA--TA-CTTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCACCTCTCxCCA--TA-CTTAGT.............. | 21 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCACCTCTCTCCA--CA-CTTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................GTCAACTCTCTCCA--TA-CTTAGT.............. | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ................................................................................................TTCACATCTCTACA--TA-CTTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ................................................................................................TTCACCTGTCTCCA--TG-CTTAGT.............. | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .................................................................................................TCACCTCTCTCCA--TA-CTTATT.............. | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .................................................................................................TCACCTTTCTCCA--TA-CTTAGTT............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............................................................................................CTTCACCTCTCTCCA--TA-CTTAG............... | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |