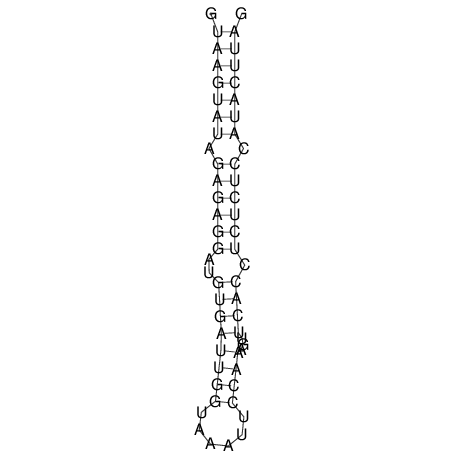
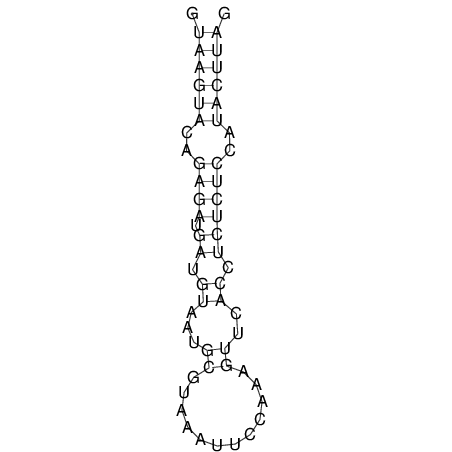
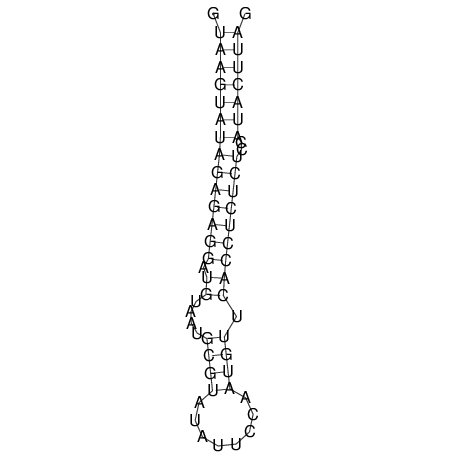
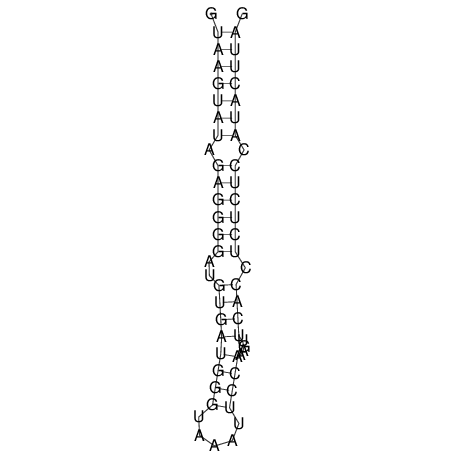
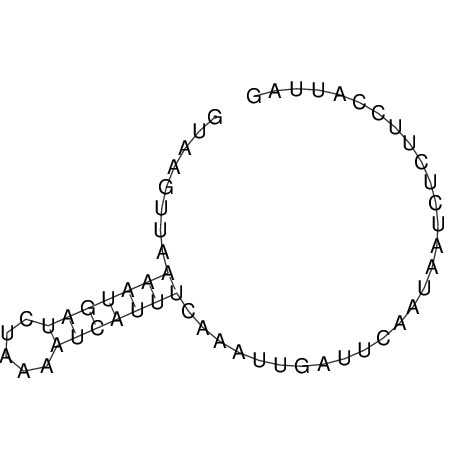
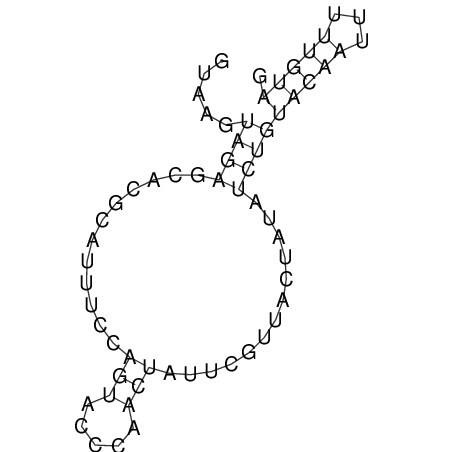
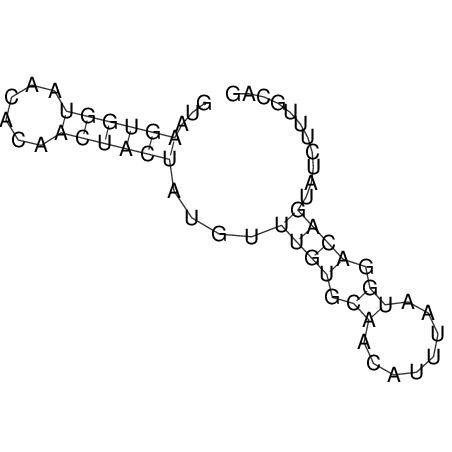
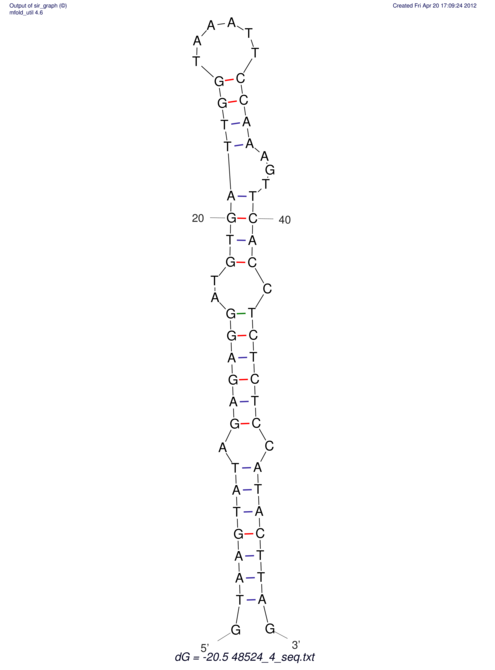
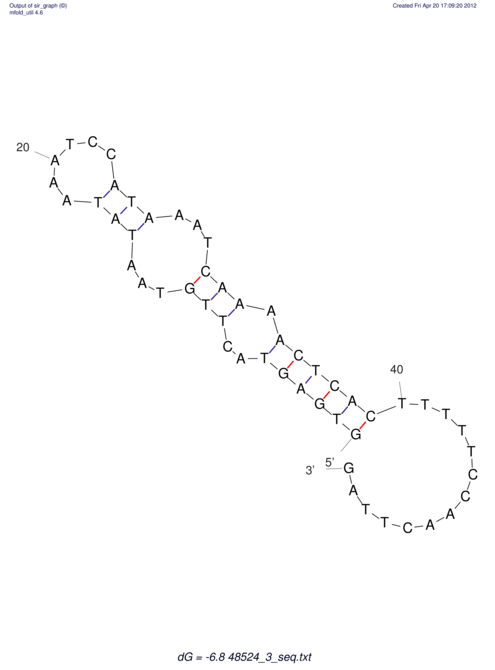
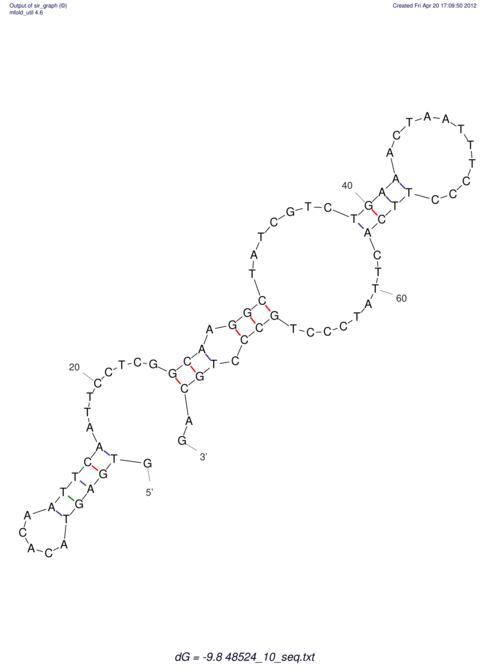
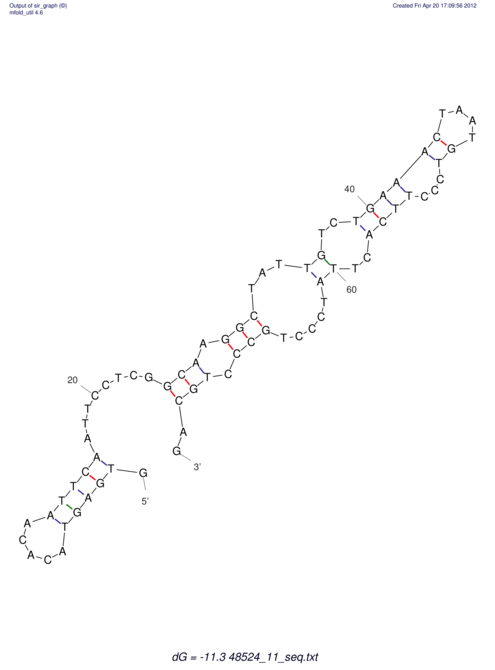
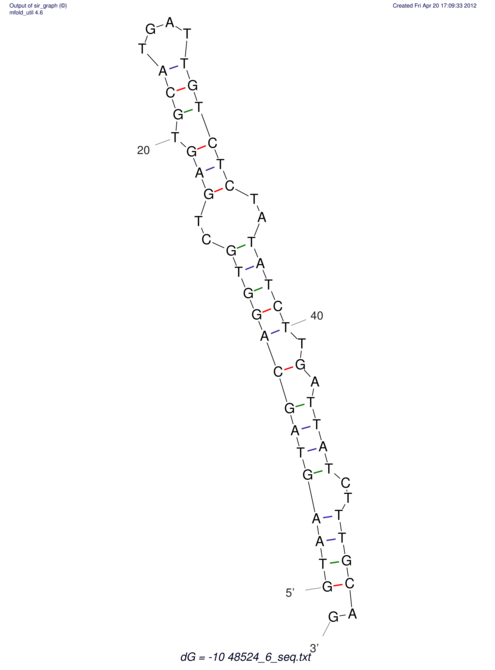
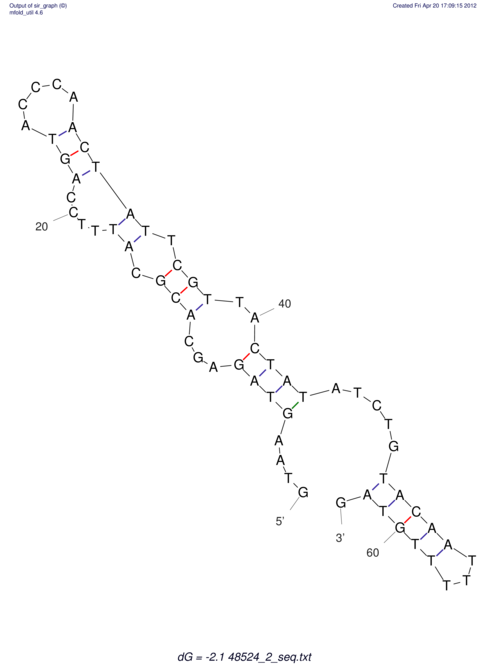
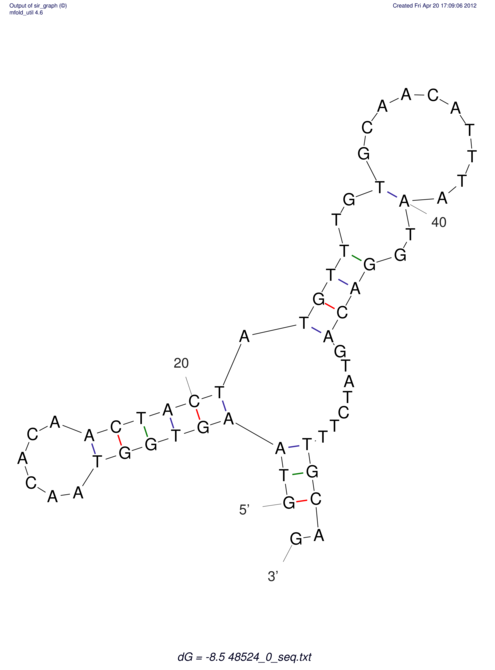
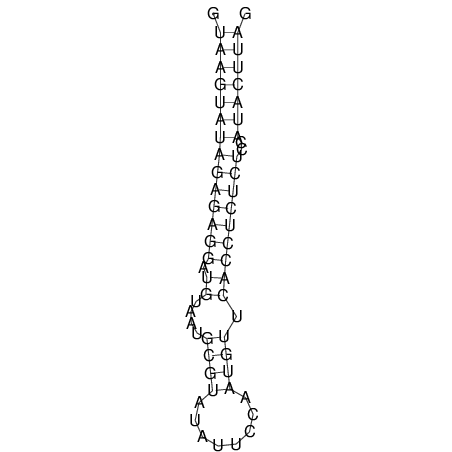
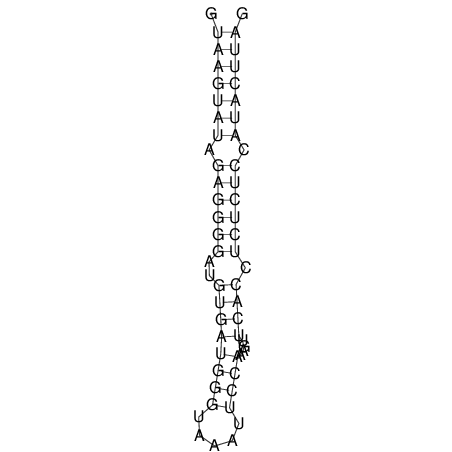
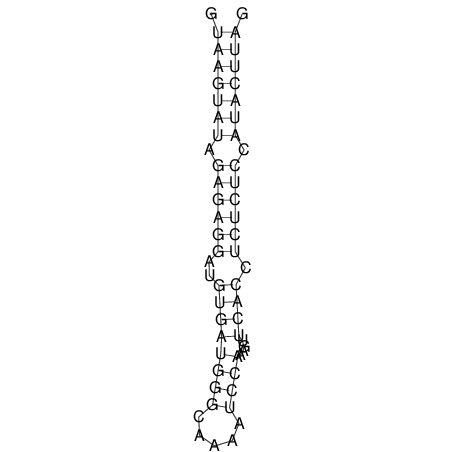
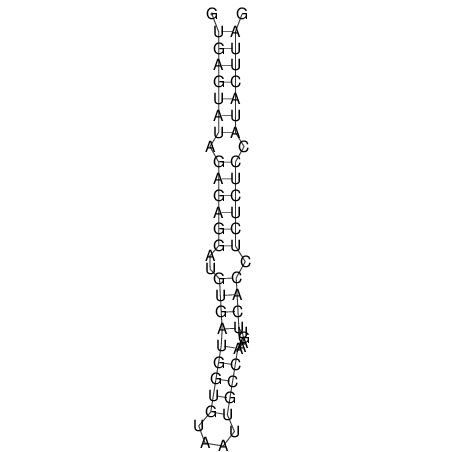
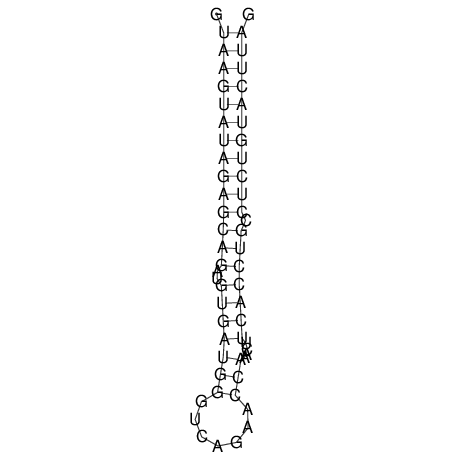
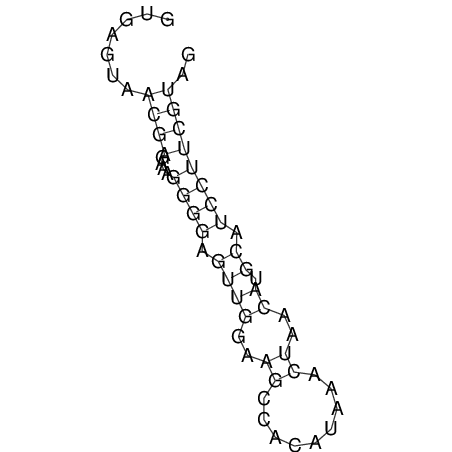
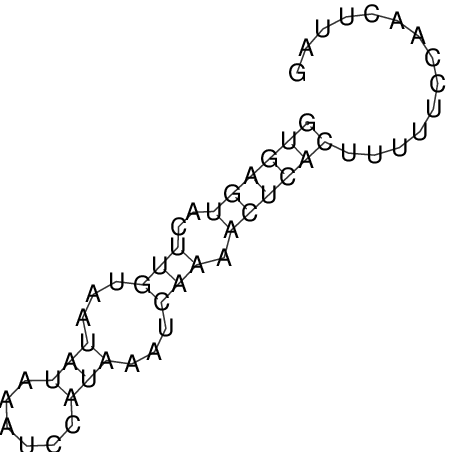
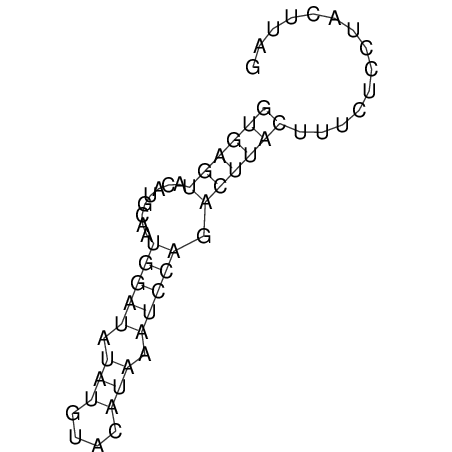
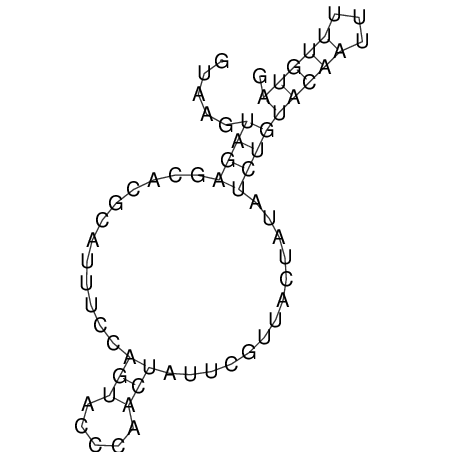
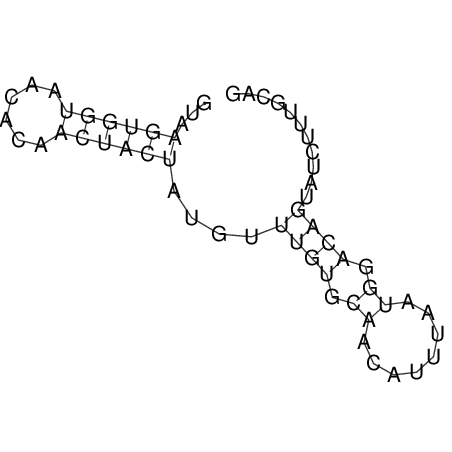
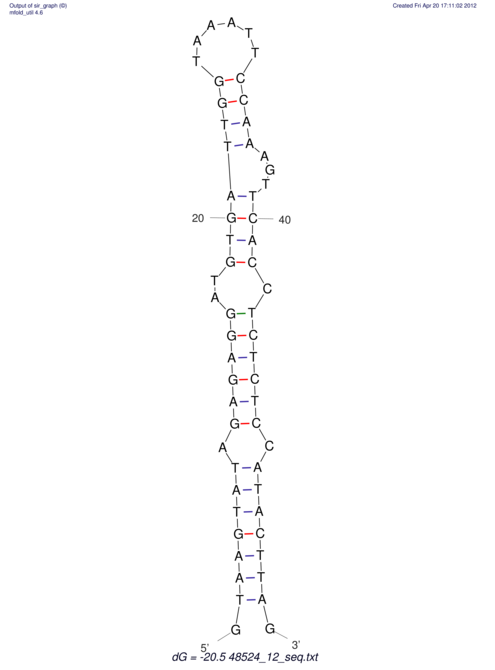
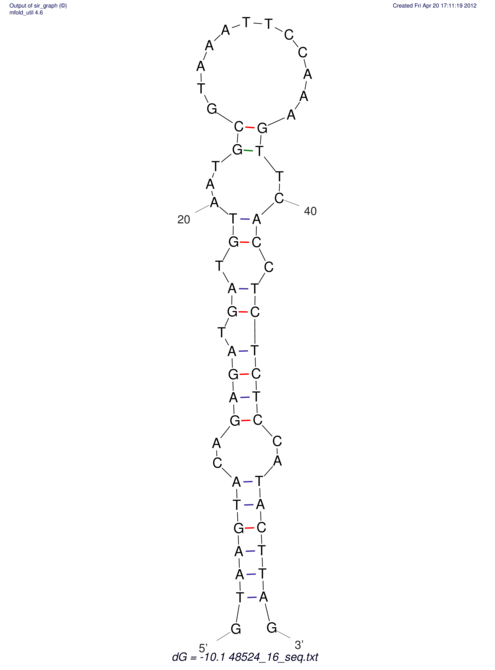
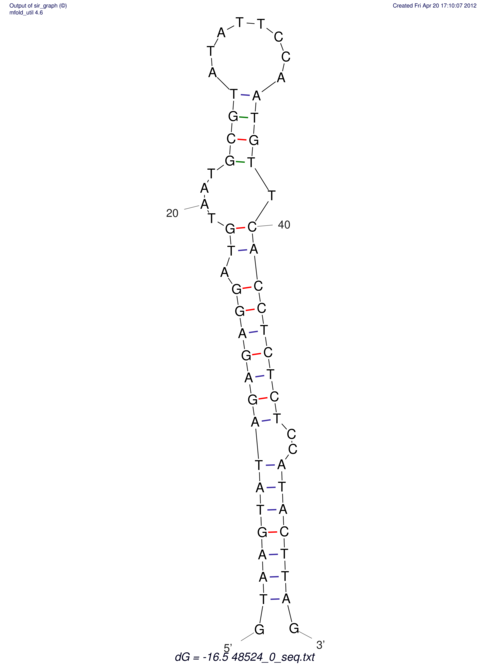
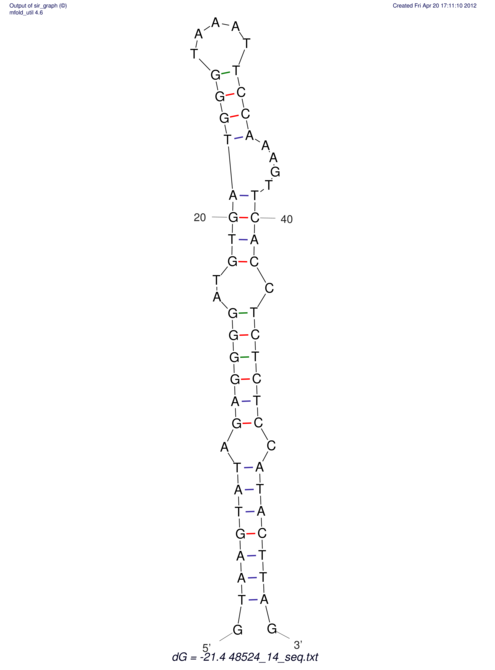
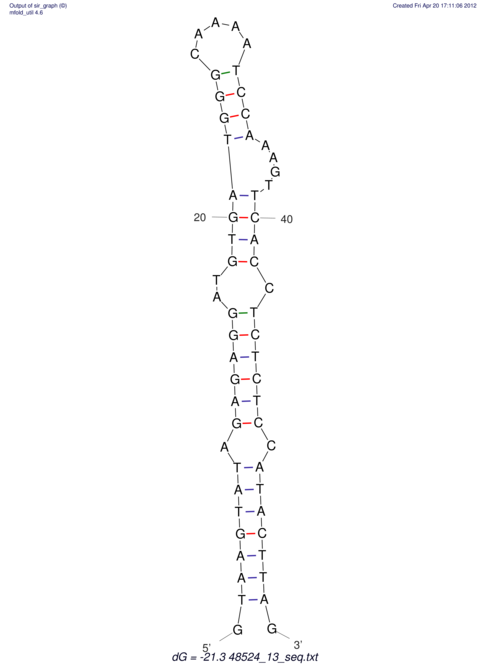
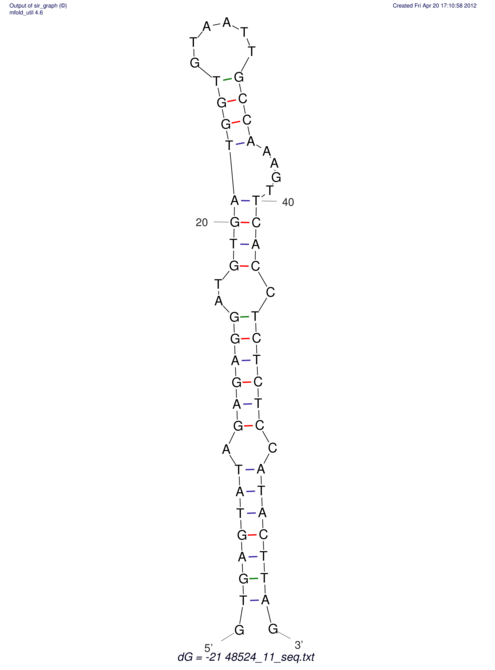
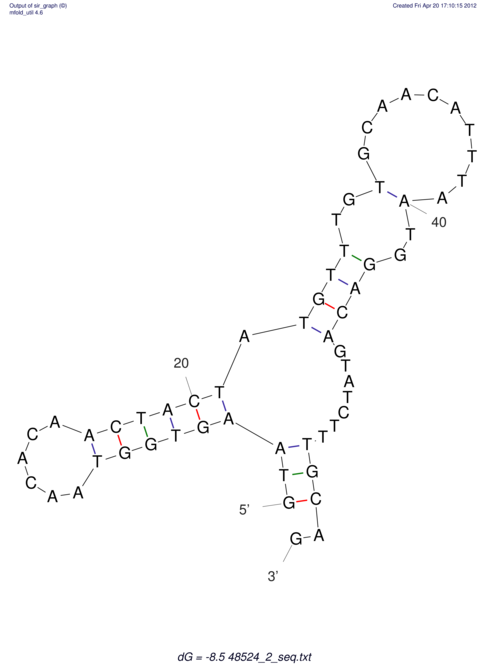
| CTCCAAGCTGTTTCAGTAAGTATAGAGAGGATGTGATGGGCAAAA------TCCAAA----------GTT-----------------CACCTCTCTCCATAC-TTAGTCGTCGAGGGGTTAT | Size | Hit Count | Total Norm | V040
Embryo | V060
Head | V047
Embryo | V108
Head |
| ....................................................................TT-----------------CACCTCTCTCCATAC-TTAGT.............. | 22 | 1 | 866.00 | 21.00 | 58.00 | 0.00 | 787.00 |
| .....................................................................T-----------------CACCTCTCTCCATAC-TTAGT.............. | 21 | 1 | 58.00 | 1.00 | 5.00 | 0.00 | 52.00 |
| .....................................................................T-----------------CACCTCTCTCCATAC-TTAGTT............. | 22 | 1 | 36.00 | 3.00 | 4.00 | 0.00 | 29.00 |
| ....................................................................TT-----------------CACCTCTCTCCATAC-TTAG............... | 21 | 1 | 22.00 | 0.00 | 1.00 | 0.00 | 21.00 |
| ...............GTAAGTATAGAGAGGATGTGATGG................................................................................... | 24 | 1 | 12.00 | 0.00 | 0.00 | 0.00 | 12.00 |
| ...............GTAAGTATAGAGAGGATGTGA...................................................................................... | 21 | 1 | 11.00 | 0.00 | 1.00 | 0.00 | 10.00 |
| ....................................................................TT-----------------CACCTCTCTCAATAC-TTAGT.............. | 22 | 1 | 9.00 | 0.00 | 0.00 | 0.00 | 9.00 |
| ...............GTAAGTATAGAGAGGATGTGATGGGC................................................................................. | 26 | 1 | 8.00 | 0.00 | 0.00 | 0.00 | 8.00 |
| ...............GTAAGTATAGAGAGGATGTGATG.................................................................................... | 23 | 1 | 7.00 | 0.00 | 1.00 | 0.00 | 6.00 |
| ....................................................................TT-----------------CACATCTCTCCATAC-TTAGT.............. | 22 | 1 | 7.00 | 0.00 | 0.00 | 0.00 | 7.00 |
| ....................................................................TT-----------------CACCTCTATCCATAC-TTAGT.............. | 22 | 1 | 6.00 | 0.00 | 0.00 | 0.00 | 6.00 |
| ....................................................................TT-----------------CACCTATCTCCATAC-TTAGT.............. | 22 | 1 | 6.00 | 0.00 | 0.00 | 0.00 | 6.00 |
| ....................................................................TT-----------------CACCTCTCTCCATAC-TTA................ | 20 | 1 | 5.00 | 0.00 | 1.00 | 0.00 | 4.00 |
| ....................................................................TT-----------------AACCTCTCTCCATAC-TTAGT.............. | 22 | 1 | 4.00 | 0.00 | 0.00 | 0.00 | 4.00 |
| ...............GTAAGTATAGAGAGGATGTGATGGG.................................................................................. | 25 | 1 | 4.00 | 1.00 | 0.00 | 0.00 | 3.00 |
| ....................................................................TT-----------------CACCTCTCTCCATAC-TT................. | 19 | 1 | 4.00 | 0.00 | 0.00 | 0.00 | 4.00 |
| ....................................................................TT-----------------CACCTCTCTACATAC-TTAGT.............. | 22 | 1 | 3.00 | 0.00 | 0.00 | 0.00 | 3.00 |
| ....................................................................TT-----------------CACCTCTCTCCATAA-TTAGT.............. | 22 | 1 | 3.00 | 0.00 | 0.00 | 0.00 | 3.00 |
| ....................................................................TT-----------------CACCTCTCTCCATAC-CTAGT.............. | 22 | 1 | 3.00 | 0.00 | 0.00 | 0.00 | 3.00 |
| ........................................................................................ACCTCTCTCCATAC-TTAGT.............. | 19 | 1 | 3.00 | 1.00 | 1.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACCTCTCTCCATAC-TTAGA.............. | 22 | 1 | 3.00 | 0.00 | 1.00 | 0.00 | 2.00 |
| ....................................................................TT-----------------CACCTCTCTCCATAC-TTATT.............. | 22 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| .....................................................................T-----------------CACCTCTCTCCATAC-TTAGTA............. | 22 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| ....................................................................TT-----------------CACCTCTTTCCATAC-TTAGT.............. | 22 | 1 | 2.00 | 0.00 | 1.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CAACTCTCTCCATAC-TTAGT.............. | 22 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| ....................................................................TT-----------------CACCTCTCTCCATAC-TTGGT.............. | 22 | 1 | 2.00 | 1.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACCCCTCTCCATAC-TTAGT.............. | 22 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| ....................................................................TT-----------------CACCTCTCTCCATAC-TTAGTT............. | 23 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| ....................................................................TT-----------------CACCTCTCTCCATCC-TTAGT.............. | 22 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| ...................................................................CTT-----------------CACCTCTCTCCATAC-TTAGT.............. | 23 | 1 | 2.00 | 0.00 | 1.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CGCCTCTCTCCATAC-TTAGT.............. | 22 | 1 | 2.00 | 0.00 | 1.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACCTCTCTCCATAC-TTAGC.............. | 22 | 1 | 2.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| ...............GTAAGTATAGAGAGGATGTGATCGGC................................................................................. | 26 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............TTAAGTATAGAGAGGATGTG....................................................................................... | 20 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACCTCTCTCTATAC-TTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACCTCTCTCCCTAC-TTAGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ................TAAGTATAGAGAGGATGTGATG.................................................................................... | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .....................................................................T-----------------CACCTCTCTCCATAC-TTAGTAA............ | 23 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTATAGAGAGGATGT........................................................................................ | 19 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ......................................................AAA----------GTT-----------------CACCTCTCTCCATAC-T.................. | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................................................T-----------------CACCTCTCTCCCTAC-TTAGT.............. | 21 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACCTCTCTCCATCC-TTCGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ...............GTAAGTATAGAGATGATGT........................................................................................ | 19 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACCTCTCTCCATAC-TTAGCA............. | 23 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTATAGAGAGGATGTG....................................................................................... | 20 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTATAGAGAGGATGTGAT..................................................................................... | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...................................................................GTT-----------------CACCTCTCTCCATAC-TTAGT.............. | 23 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CAACTCTCTCAATAC-TT................. | 19 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACCTCTCTACATAA-TTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACACCTCTCCATAC-TTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................CT-----------------CACCTCTCTCCATAC-TTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .................AAGTATAGAGAGGATGTGATGGGC................................................................................. | 24 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .......................................................................................CACCTCTCTCCATAC-TTAGT.............. | 20 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .....................................................................T-----------------CACCTCTCTCCATAA-TTAGT.............. | 21 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .....................................................................T-----------------AACCTCTCTCCATAC-TTAGT.............. | 21 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .....................................................................T-----------------CACCTCTCTCCATAC-TTAGTGT............ | 23 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACCTCTCTCCATAC-TTAGG.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .....................................................................T-----------------CACCTCTCTCCATAC-TTAG............... | 20 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------GACCTCTCTCCATAC-TTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTATAGAGAGGATTTGATGGGC................................................................................. | 26 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............TTAAGTATAGAGAGGATGCGA...................................................................................... | 21 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CAGCTCTCTCCATAC-TTAGT.............. | 22 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| .....................................................................T-----------------CACATCTCTCCATAC-TTAGTT............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTATAGAGAGGATGTGATC.................................................................................... | 23 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...............GTAAGTATAGAGAGGATGTCAT..................................................................................... | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .....................................................................T-----------------CACCTATCTCCATAC-TTAGT.............. | 21 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .........................................................................................CCTCTCTCCATAC-TTAGT.............. | 18 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| .....................................................................T-----------------CACCTCTCTCCATAC-TTAGAT............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACCTCTCTGCATAC-TTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACCTCTCxCCATAC-TTAGT.............. | 21 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACCTCTCTCCACAC-TTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................GT-----------------CAACTCTCTCCATAC-TTAGT.............. | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ....................................................................TT-----------------CACATCTCTACATAC-TTAGT.............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ....................................................................TT-----------------CACCTGTCTCCATGC-TTAGT.............. | 22 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................................................T-----------------CACCTCTCTCCATAC-TTATT.............. | 21 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .....................................................................T-----------------CACCTTTCTCCATAC-TTAGTT............. | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ...................................................................CTT-----------------CACCTCTCTCCATAC-TTAG............... | 22 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 |