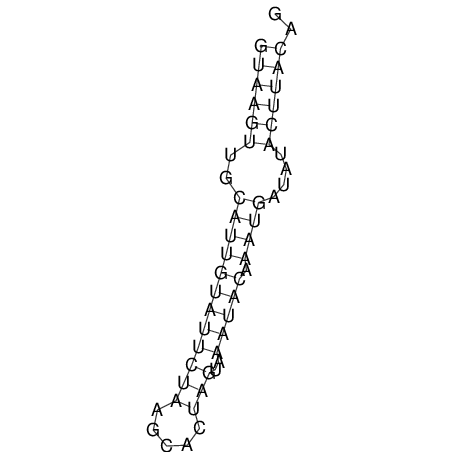
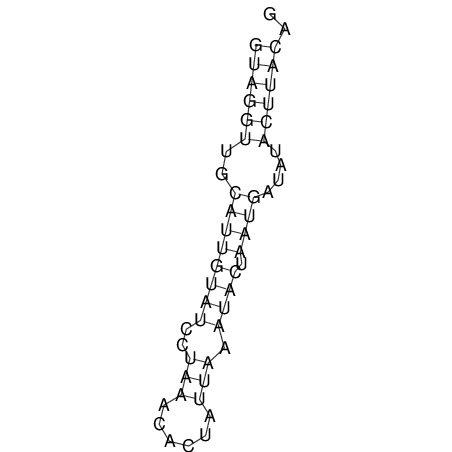
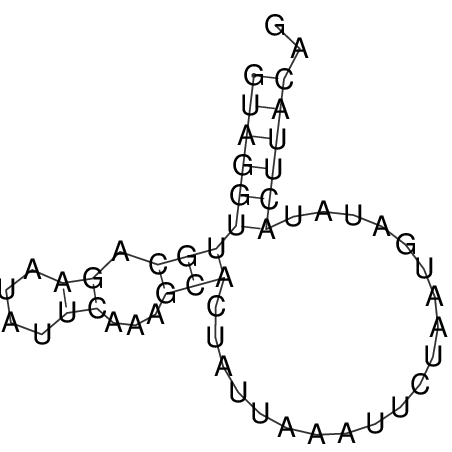
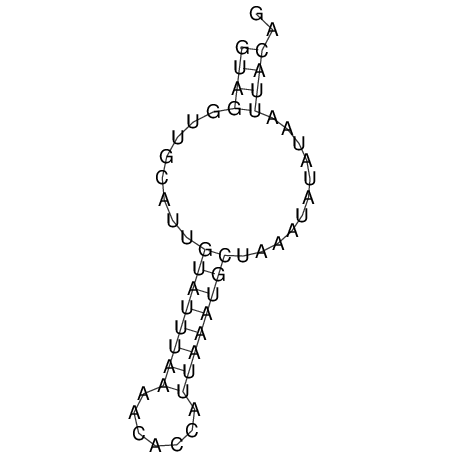
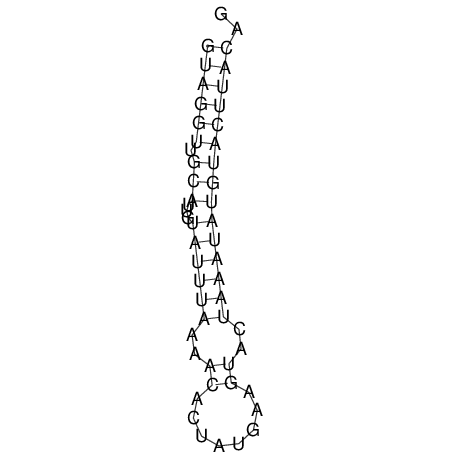

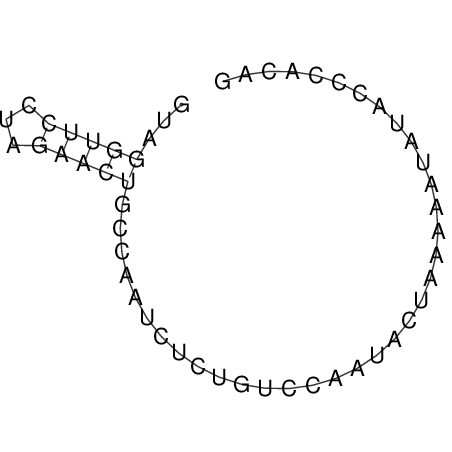
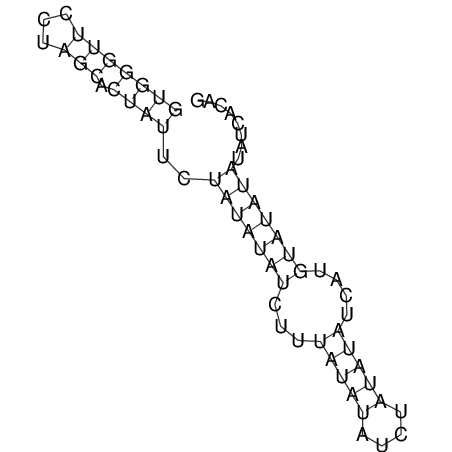
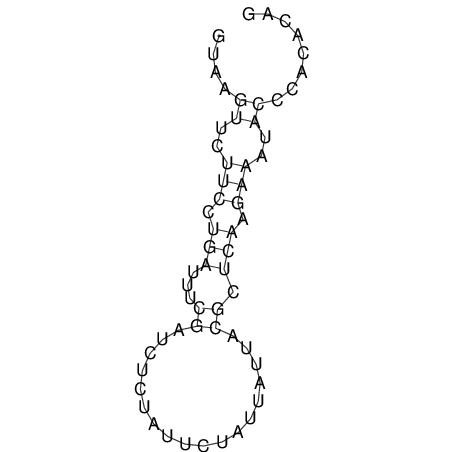
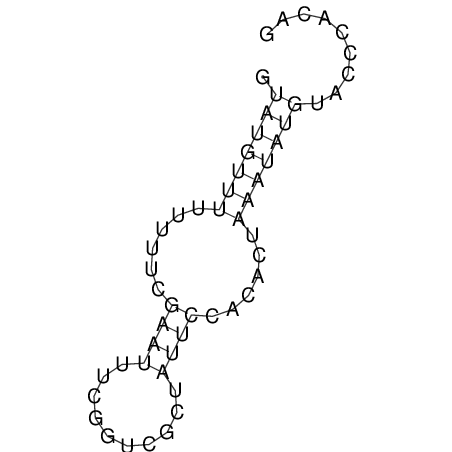
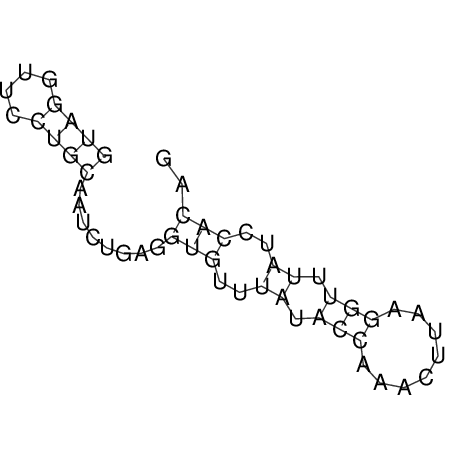
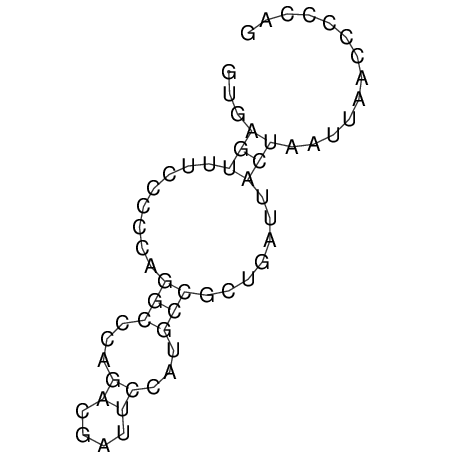
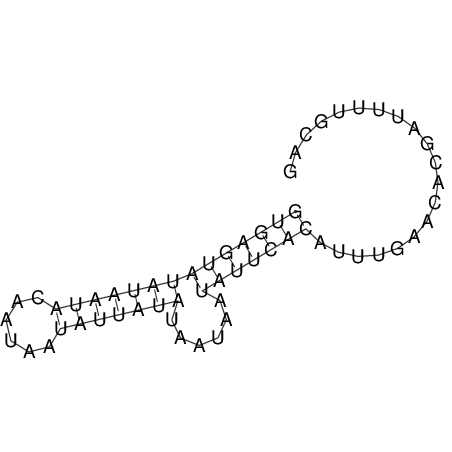
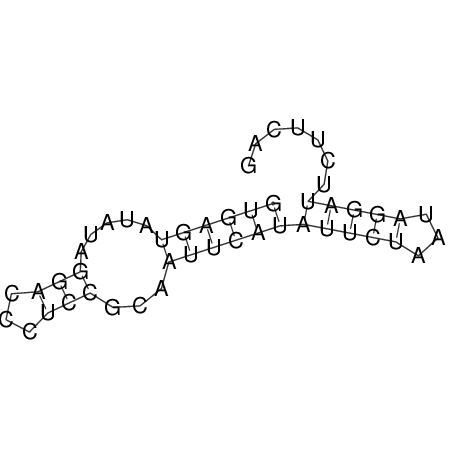
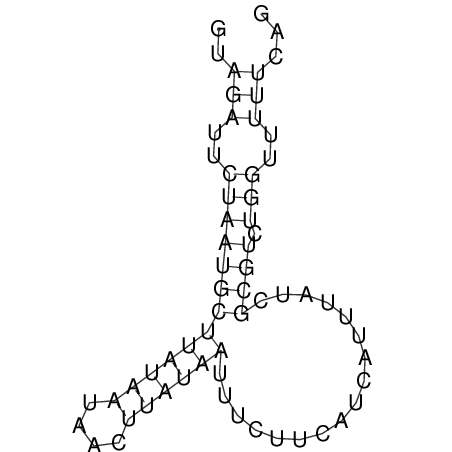

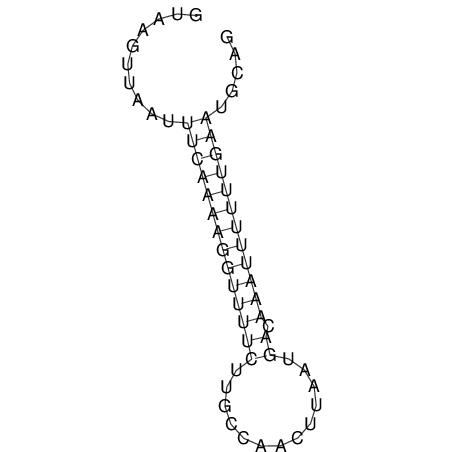
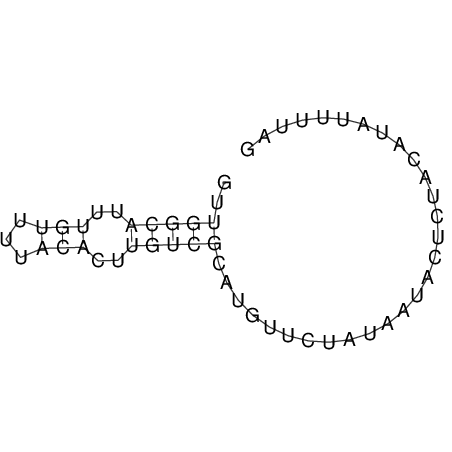
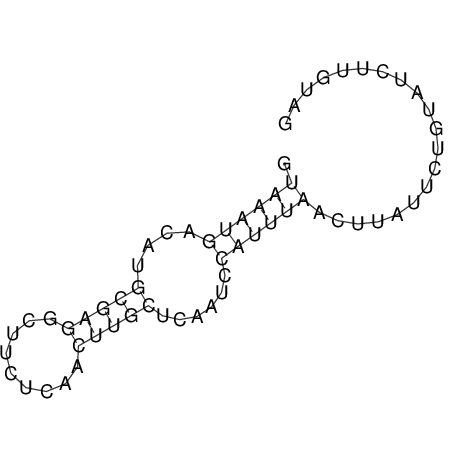

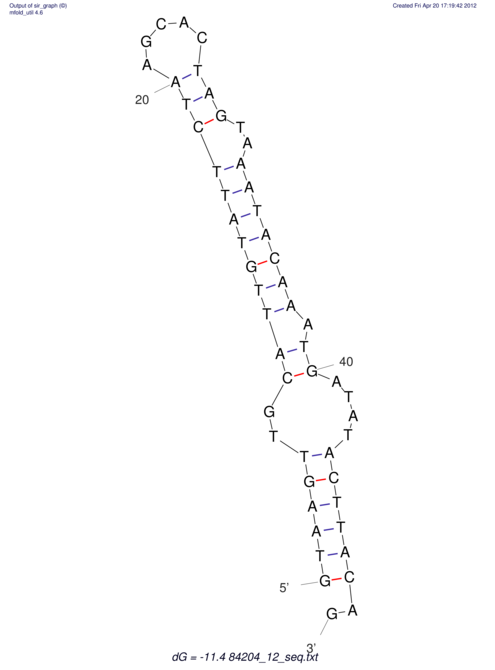
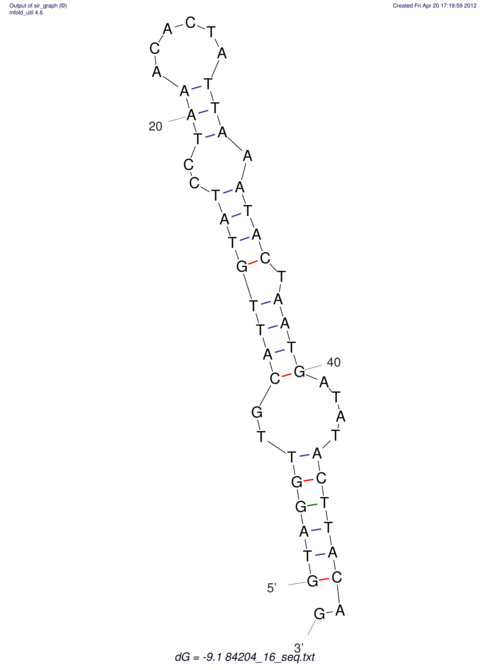

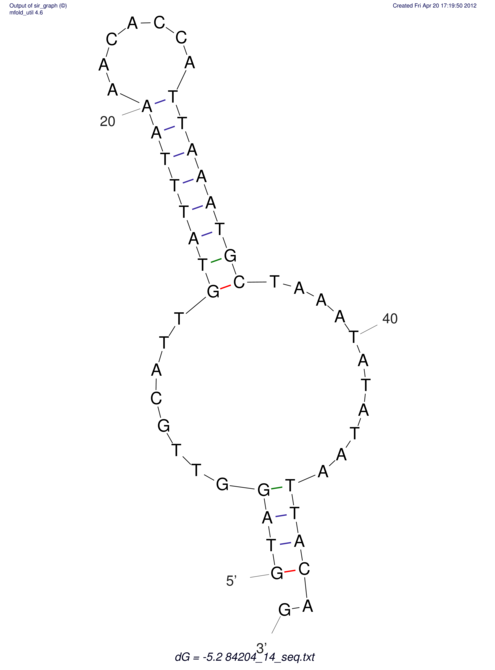

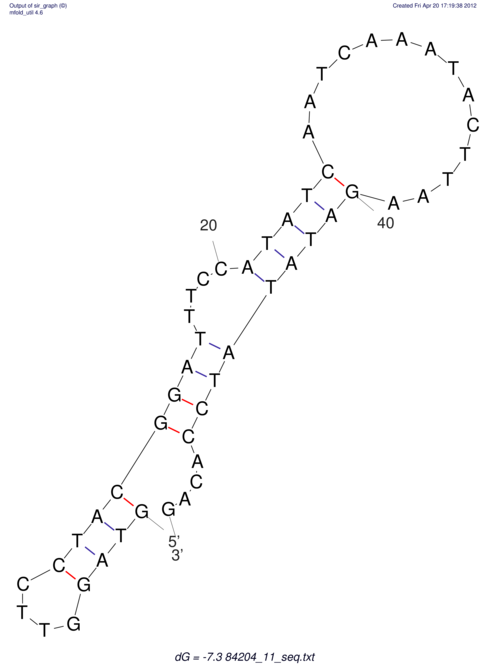
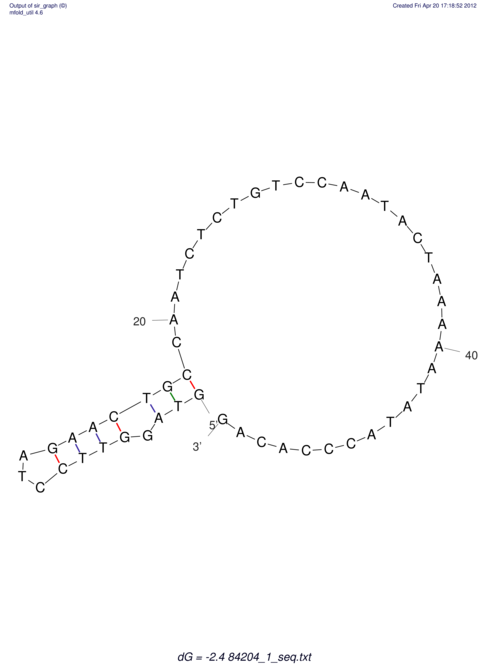
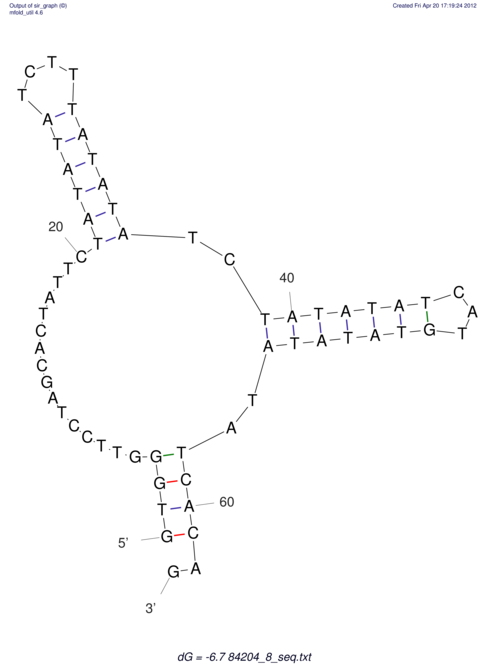
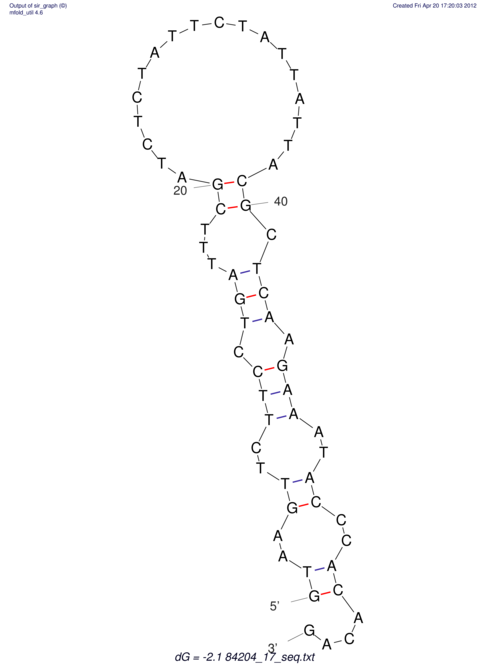



View alignment of MAF (12-files) vs LASTZ top hits (20-flies)
View all possible LASTZ hits (20-flies)
CG3160_in3 |
chrX:5582203-5582254 - |
Candidate |
|---|---|---|
| [View on UCSC Genome Browser {Cornell Mirror}] | mirtron |
Legend:
mature strand at position 5582230-5582254| Species | Coordinate | Source | Confidence | Alignment |
| dm3 | chrX:5582188-5582269 - | 1 | TTGTCTACCTTTGTGGTAAGTTGCAT--TGTATTCTAA---------------------------GCACTAGT--AAAT------------------ACAAATG--A-------------------------TATACTTACAGAAGCCTCGAAGGCGT | |
| droSim1 | chrX:4333816-4333897 - | LASTZ | 4 | TTGTCCACCTTTGTGGTAGGTTGCAT--TGTATCCTAA---------------------------ACACTATT--AAAT------------------ACTAATG--A-------------------------TATACTTACAGAAGCCCCGAAGGCGT |
| droSec1 | super_4:1176184-1176265 + | LASTZ | 5 | TTGTCCACCTTTGTGGTAGGTTGCAG--AATATTCAAA---------------------------GCACTATT--AAAT------------------TCTAATG--A-------------------------TATACTTACAGAAGCCCCGAAGGCGT |
| droYak2 | chrX:2886884-2886965 + | LASTZ | 5 | TTGTCCACCTTTGTGGTAGGTTGCAT--TGTATTTAAA---------------------------ACACCATT--AAAT------------------GCTAAAT--A-------------------------TATAATTACAGAAGCCTCGAAGGCGT |
| droEre2 | scaffold_4690:2935043-2935124 - | LASTZ | 4 | TTGTCAACCTTTGTGGTAGGTTGCAT--TGTATTTAAA---------------------------ACACTATG--AAGT------------------ACTAAAT--A-------------------------TGTACTTACAGAAGCCTCGAAGGCGT |
| droEug1 | scf7180000409095:622922-623003 - | LASTZ | 5 | TTATCCACATTTGTAGTAGGTTCCTA--CGGATTTCCA---------------------------TATCAATC--AAAT------------------ACTTAAG--A-------------------------TATATCCACAGAAACCACGGAGACGA |
| droBia1 | scf7180000302069:240686-240767 + | LASTZ | 5 | CTAGCCACCTTTGTCGTAGGTTCCTA--GAACTGCCAA---------------------------TCTCTGTC--CAAT------------------ACTAAAA--A-------------------------TATACCCACAGAAGCCAAGAAGACGA |
| droTak1 | scf7180000415704:943029-943121 + | LASTZ | 5 | TTAGCCACCTTTGTCGTGGGTTCCTA--GCACTATTCT---ATAT-----------ATCT-TT-ATATATCTATAT--ATC--------------AT----G-TA-T-------------------------ATATATCACAGAAACCGCGGAGACGG |
| droEle1 | scf7180000491001:865333-865422 + | LASTZ | 5 | CTGTCCACCTTTGTGGTAAGTTCTTC--CTGATTTCGA---------------------------TCTCTATT--CTAT------TATTAC------GCTCAAGAAA-------------------------TACCCACACAGAAGTCACGAAGGCGT |
| droRho1 | scf7180000779976:691191-691274 + | LASTZ | 5 | TTGTCTACATTTGTGGTATGTTTTTTTTCGAATTTCGG---------------------------TCGCTATT--CCAC------------------ACTAAAT--A-------------------------TGTACCCACAGAAGCCGCGAAGCCGT |
| droFic1 | scf7180000454072:2528858-2528939 + | LASTZ | 5 | CTGTCGACTTTTGTGGTAGGTTCCTG--CAATCTGAGG---------------------------TGTTTATA--CCAA------------------ACTTAAG--G-------------------------TTTATCCACAGAAGCCACGCAGGCGT |
| droKik1 | scf7180000302344:685348-685434 - | LASTZ | 5 | CTCGCCCACTTTGTGGTGAGTTTCCC--CCAGGCCCAG---------------------------ACG--ATT--CCAT------GCCGCT------GATTACTA-A-------------------------TTAACCCCCAGAAACCACGAAGGCGT |
| droAna3 | scaffold_13117:3260782-3260871 - | LASTZ | 5 | CTGCACACCTTTGTGGTGAGTATATA--ATACAATAAT---------------------------ATTATATA--ATAA------TATTCAC---A-TTTGAAC--A-------------------------CGATTTTGCAGAAACCACGAAGGCGC |
| droBip1 | scf7180000396434:597307-597385 + | LASTZ | 5 | CTCCACAGCTTTGTGGTGAGTATATA--GGACCCT-CC---------------------------GCAAT-TC--ATAT------------------TCTAA-T--A-------------------------GGATTCTTCAGAAACCACGCAGGCGC |
| dp4 | chrXL_group1e:588994-589086 - | LASTZ | 5 | CTGTCTCACTTTGTGGTAGATTCTAA--TGCTTATAATAACTTATAATTTCTTCATCA---------------TTT--ATC--------------GC----G-T--C-------------------------TGGTTTTTCAGAAGCGGCCAAGTCGC |
| droPer1 | super_30:621799-621888 + | LASTZ | 5 | CTGTCTCACTTTGTGGTAGATTCTAA--TGCTTATAAC---TTATAATTTCTTCATCA---------------TTT--ATC--------------GC----G-T--C-------------------------TGGTTTTTCAGAAGCGGCCAAGTCGC |
| droWil1 | scaffold_180702:827721-827807 - | LASTZ | 5 | ATGGCACATTTTATCGTAAGTTAATT--TCAAAA---G-----------------GTT--TTCTTGCCAACTT--AATG------------------ACAAATT--T-------------------------TTTGAATGCAGAAACCCAGGAGTCGC |
| droVir3 | scaffold_12928:357066-357149 - | LASTZ | 5 | GTGGCGCATTTTGTGGTTGGCATTTG--TTT-----------------------TACA---------------CTT--GTCGCATGTTCTATAAT--ACTCTAC--A----------------------------TATTTTAGAAACCGAGGATACGG |
| droMoj3 | scaffold_6328:487653-487744 + | LASTZ | 5 | TTGGGACACTTTGTGGTAAATGACAT--GCGAGGCTTC----------------TCAACT-TG-CTCAATCCATTT--A------------------ACTTATTC-T-------------------------GTATCTTGTAGAAATCGCCAACACGT |
| droGri2 | scaffold_15203:6798783-6798893 - | LASTZ | 5 | CTGTCGCACTTTGTGGTAAGCAAACA--AGCA-----------------------ATA--TT---ATT-------------GTGTGATTAAAAAA--ACGAAAT--ATTGTTACTATCAAATCTTCGTTTTCTATCGATCTAGAAACAACGAACTCGC |
| Species | Alignment |
| dm3 | TTGTCTACCTTTGTGGTAAGTTGCAT--TGTATTCTAA---------------------------GCACTAGT--AAAT------------------ACAAATG--A-------------------------TATACTTACAGAAGCCTCGAAGGCGT |
| droSim1 | TTGTCCACCTTTGTGGTAGGTTGCAT--TGTATCCTAA---------------------------ACACTATT--AAAT------------------ACTAATG--A-------------------------TATACTTACAGAAGCCCCGAAGGCGT |
| droSec1 | TTGTCCACCTTTGTGGTAGGTTGCAG--AATATTCAAA---------------------------GCACTATT--AAAT------------------TCTAATG--A-------------------------TATACTTACAGAAGCCCCGAAGGCGT |
| droYak2 | TTGTCCACCTTTGTGGTAGGTTGCAT--TGTATTTAAA---------------------------ACACCATT--AAAT------------------GCTAAAT--A-------------------------TATAATTACAGAAGCCTCGAAGGCGT |
| droEre2 | TTGTCAACCTTTGTGGTAGGTTGCAT--TGTATTTAAA---------------------------ACACTATG--AAGT------------------ACTAAAT--A-------------------------TGTACTTACAGAAGCCTCGAAGGCGT |
| droEug1 | TTATCCACATTTGTAGTAGGTTCCTA--CGGATTTCCA---------------------------TATCAATC--AAAT------------------ACTTAAG--A-------------------------TATATCCACAGAAACCACGGAGACGA |
| droBia1 | CTAGCCACCTTTGTCGTAGGTTCCTA--GAACTGCCAA---------------------------TCTCTGTC--CAAT------------------ACTAAAA--A-------------------------TATACCCACAGAAGCCAAGAAGACGA |
| droTak1 | TTAGCCACCTTTGTCGTGGGTTCCTA--GCACTATTCT---ATAT-----------ATCT-TT-ATATATCTATAT--ATC--------------AT----G-TA-T-------------------------ATATATCACAGAAACCGCGGAGACGG |
| droEle1 | CTGTCCACCTTTGTGGTAAGTTCTTC--CTGATTTCGA---------------------------TCTCTATT--CTAT------TATTAC------GCTCAAGAAA-------------------------TACCCACACAGAAGTCACGAAGGCGT |
| droRho1 | TTGTCTACATTTGTGGTATGTTTTTTTTCGAATTTCGG---------------------------TCGCTATT--CCAC------------------ACTAAAT--A-------------------------TGTACCCACAGAAGCCGCGAAGCCGT |
| droFic1 | CTGTCGACTTTTGTGGTAGGTTCCTG--CAATCTGAGG---------------------------TGTTTATA--CCAA------------------ACTTAAG--G-------------------------TTTATCCACAGAAGCCACGCAGGCGT |
| droKik1 | CTCGCCCACTTTGTGGTGAGTTTCCC--CCAGGCCCAG---------------------------ACG--ATT--CCAT------GCCGCT------GATTACTA-A-------------------------TTAACCCCCAGAAACCACGAAGGCGT |
| droAna3 | CTGCACACCTTTGTGGTGAGTATATA--ATACAATAAT---------------------------ATTATATA--ATAA------TATTCAC---A-TTTGAAC--A-------------------------CGATTTTGCAGAAACCACGAAGGCGC |
| droBip1 | CTCCACAGCTTTGTGGTGAGTATATA--GGACCCT-CC---------------------------GCAAT-TC--ATAT------------------TCTAA-T--A-------------------------GGATTCTTCAGAAACCACGCAGGCGC |
| dp4 | CTGTCTCACTTTGTGGTAGATTCTAA--TGCTTATAATAACTTATAATTTCTTCATCA---------------TTT--ATC--------------GC----G-T--C-------------------------TGGTTTTTCAGAAGCGGCCAAGTCGC |
| droPer1 | CTGTCTCACTTTGTGGTAGATTCTAA--TGCTTATAAC---TTATAATTTCTTCATCA---------------TTT--ATC--------------GC----G-T--C-------------------------TGGTTTTTCAGAAGCGGCCAAGTCGC |
| droWil1 | ATGGCACATTTTATCGTAAGTTAATT--TCAAAA---G-----------------GTT--TTCTTGCCAACTT--AATG------------------ACAAATT--T-------------------------TTTGAATGCAGAAACCCAGGAGTCGC |
| droVir3 | GTGGCGCATTTTGTGGTTGGCATTTG--TTT-----------------------TACA---------------CTT--GTCGCATGTTCTATAAT--ACTCTAC--A----------------------------TATTTTAGAAACCGAGGATACGG |
| droMoj3 | TTGGGACACTTTGTGGTAAATGACAT--GCGAGGCTTC----------------TCAACT-TG-CTCAATCCATTT--A------------------ACTTATTC-T-------------------------GTATCTTGTAGAAATCGCCAACACGT |
| droGri2 | CTGTCGCACTTTGTGGTAAGCAAACA--AGCA-----------------------ATA--TT---ATT-------------GTGTGATTAAAAAA--ACGAAAT--ATTGTTACTATCAAATCTTCGTTTTCTATCGATCTAGAAACAACGAACTCGC |
| dm3 | droSim1 | droSec1 | droYak2 | droEre2 | droEug1 | droBia1 | droTak1 | droEle1 | droRho1 | droFic1 | droKik1 | droAna3 | droBip1 | dp4 | droPer1 | droWil1 | droVir3 | droMoj3 | droGri2 | |
| RNAfold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
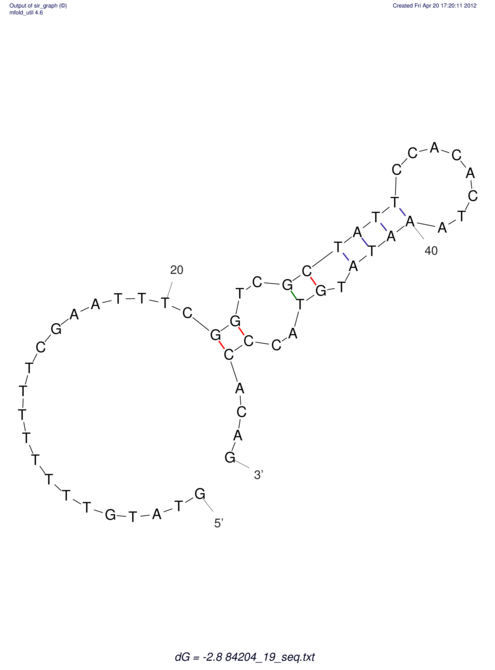
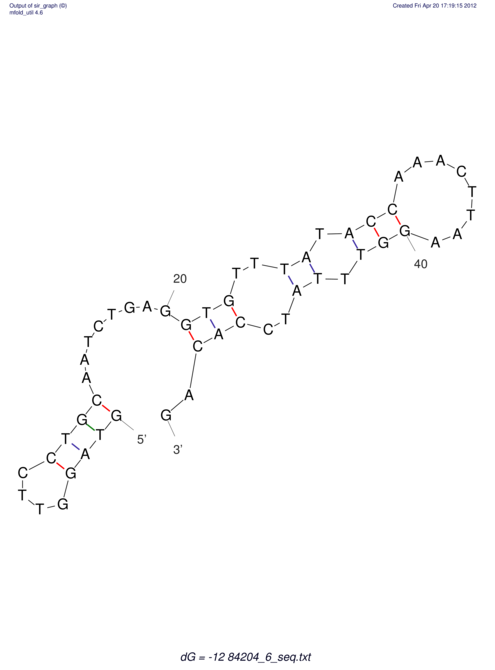
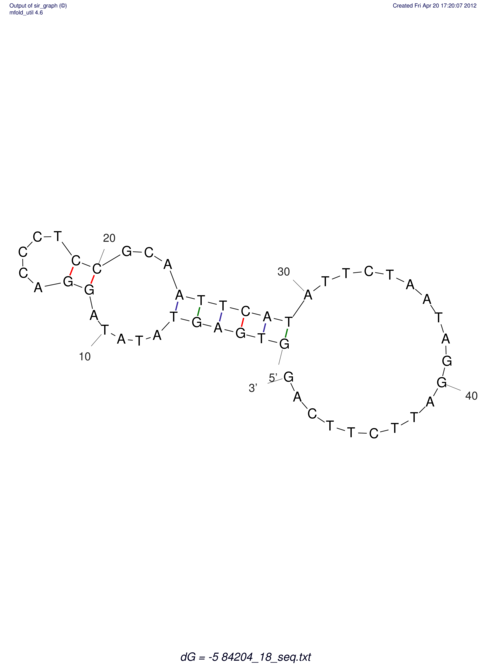
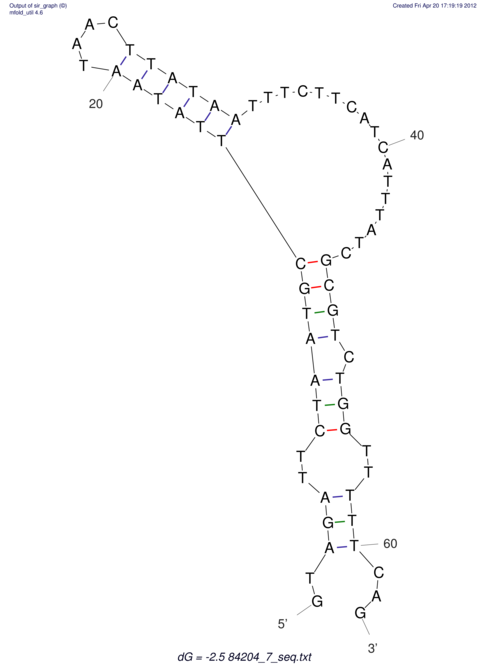
| UNAFold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Species | Alignment | ||||||||||||||||||||||||||||||||||||||||||||||||
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droSec1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droYak2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droPer1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droWil1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 04/20/2012 at 05:20:16 EDT