| (1) AGO.mut | (2) AGO1.ip | (4) AGO2.ip | (1) AGO3.ip | (10) B-CELL | (10) BRAIN | (1) CELL-LINE | (12) EMBRYO | (1) ESC | (3) FIBROBLAST | (1) HEART | (2) KIDNEY | (10) LIVER | (2) LUNG | (4) LYMPH | (15) OTHER | (1) OTHER.mut | (1) OVARY | (1) PANCREAS | (1) PIWI.ip | (1) SKIN | (9) SPLEEN | (7) TESTES | (3) THYMUS | (2) TOTAL-RNA | (1) UTERUS |
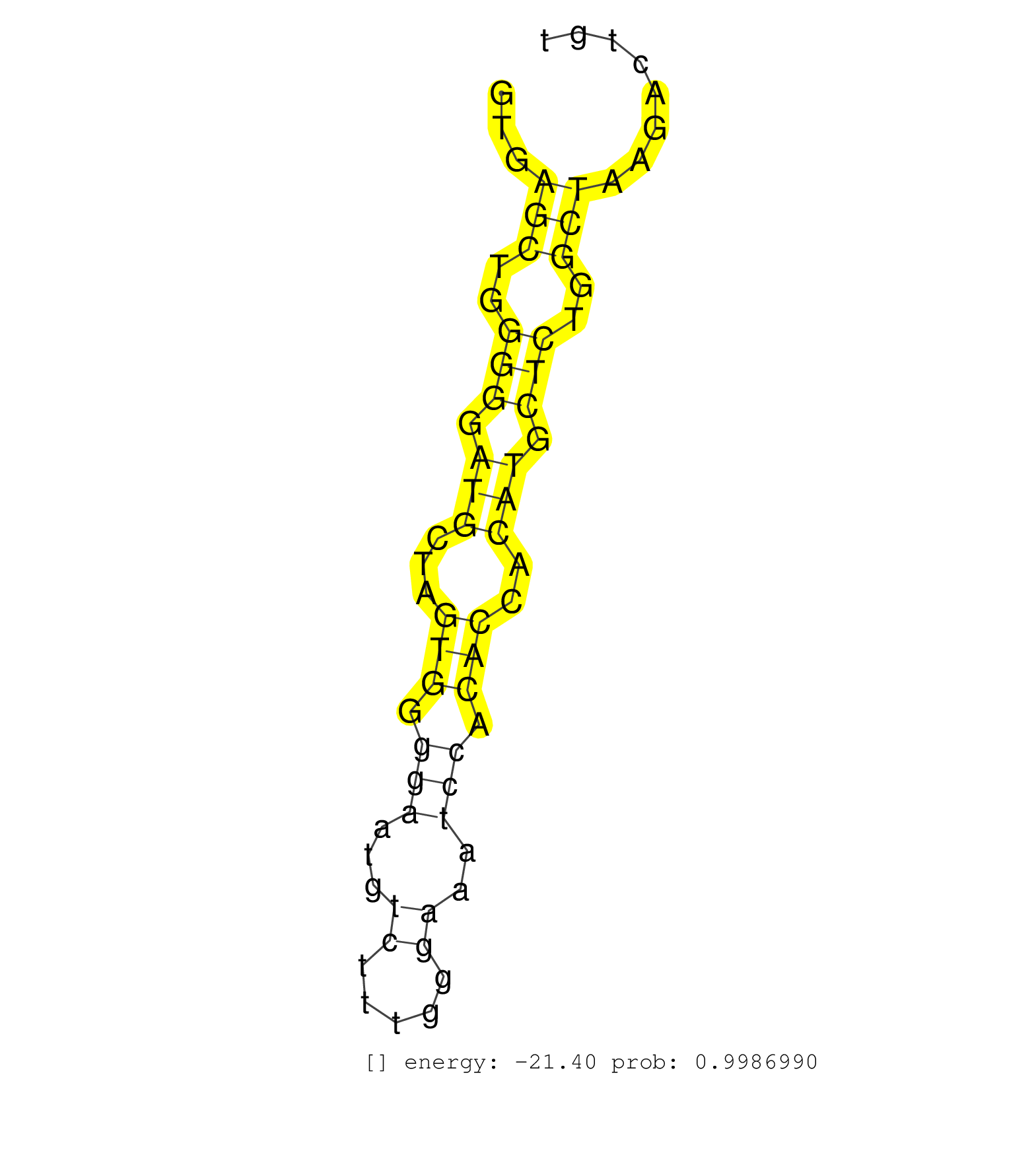
| TGTTTGGAACTGATCTTCAAGAGAAACTGAGTGACCAACACTACCTCTTGGTGAGCTGGGGGATGCTAGTGGGGAATGTCTTTGGGAAATCCACACCACATGCTCTGGCTAAGACTGTTTCTGTCACCAGGTGGGTGCTGGTGCCATTGGCTGTGAGATGCTCAAAGTCTTTGCCCTAGT .....................................................(((..(((.(((...(((.(((...((.....))..))).)))..))).)))..)))...................................................................... ..................................................51.................................................................118............................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR073955(GSM629281) total RNA. (blood) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR037905(GSM510441) brain_rep2. (brain) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | mjTestesWT3() Testes Data. (testes) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | GSM510444(GSM510444) brain_rep5. (brain) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | mjLiverWT3() Liver Data. (liver) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (salivary gland) | SRR525244(SRA056111/SRX170320) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR037936(GSM510474) 293cand1. (cell line) | SRR525240(SRA056111/SRX170316) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR525241(SRA056111/SRX170317) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR037923(GSM510461) e9p5_rep1. (embryo) | SRR059765(GSM562827) DN3_control. (thymus) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR037901(GSM510437) testes_rep2. (testes) | SRR042478(GSM539870) mouse lung tissue [09-002]. (lung) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR065058(SRR065058) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR525243(SRA056111/SRX170319) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR037929(GSM510467) e7p5_rep3. (embryo) | GSM475281(GSM475281) total RNA. (testes) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR037904(GSM510440) brain_rep1. (brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................ACACCACATGCTCTGGCTAAGA.................................................................. | 22 | 1 | 70.00 | 70.00 | - | 5.00 | 5.00 | - | 3.00 | 3.00 | 1.00 | 2.00 | - | 3.00 | 1.00 | - | 4.00 | 1.00 | 1.00 | 1.00 | 4.00 | - | - | - | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | 1.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | 2.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 |
| ............................................................................................ACACCACATGCTCTGGCTAAG................................................................... | 21 | 1 | 36.00 | 36.00 | - | 3.00 | 2.00 | - | - | 1.00 | 2.00 | - | - | - | 2.00 | 1.00 | - | - | 2.00 | 1.00 | - | 3.00 | - | - | 1.00 | - | - | 2.00 | - | - | 1.00 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................ACACCACATGCTCTGGCTAAGT.................................................................. | 22 | 1 | 19.00 | 36.00 | - | - | - | - | 2.00 | - | - | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCTAA.................................................................... | 20 | 1 | 15.00 | 15.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 2.00 | - | 1.00 | - | - | - | - | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGGGGGATGCTAGTGGGGAATGTCTTTGGGAAATCCACACCACATGCTCTGGCTAAGACTGTTTCTGTCACCAG.................................................. | 80 | 1 | 14.00 | 14.00 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCT...................................................................... | 18 | 1 | 9.00 | 9.00 | - | - | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCTATGT.................................................................. | 22 | 1 | 6.00 | 1.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CACACCACATGCTCTGGCTAAGA.................................................................. | 23 | 1 | 5.00 | 5.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CTCTGGCTAAGACTGTTTCTG......................................................... | 21 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................TACCTCTTGGTGAGCCGGC........................................................................................................................ | 19 | 4.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGAGCTGGGGGATGCTAGTGG............................................................................................................ | 22 | 1 | 4.00 | 4.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CACCACATGCTCTGGCTAAGA.................................................................. | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CACACCACATGCTCTGGCTAAG................................................................... | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCTAAGAC................................................................. | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGGGGGATGCTAGTGGGGAATGTCTTTGGGAAATCCACACCACATGCTCTGGCTAAGACTGTTTCTGTCACCAGA................................................. | 81 | 1 | 3.00 | 14.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGCTAGTGGGGAATGTCTTTGGG.............................................................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCTAAGAT................................................................. | 23 | 1 | 2.00 | 70.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCTAAT................................................................... | 21 | 1 | 2.00 | 15.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCTAT.................................................................... | 20 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGGGGGATGCTAGTGGGGAATGTCTTTGGGAAATCCACACCACATGCTCTGGCTAAGACTGTTTCTGTCACC.................................................... | 78 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................TGGGGGATGCTAGTGGGGA......................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CACACCACATGCTCTGGCTAT.................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGAGCTGGGGGATGCTAGTGA............................................................................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................CACACCACATGCTCTGGCTAA.................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCTGT.................................................................... | 20 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCTACG................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGGGGGATGCTAGTGGT........................................................................................................... | 23 | 1 | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCA...................................................................... | 18 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................CTGGCTAAGACTGTTTCTGTCA...................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................GGGAATGTCTTTGGGATG........................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................ACACCACATGCTCTGGCTAAGAAA................................................................ | 24 | 1 | 1.00 | 70.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCTAAGAA................................................................. | 23 | 1 | 1.00 | 70.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCTA..................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCTAATA.................................................................. | 22 | 1 | 1.00 | 15.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTGGCTGTGAGATGCCATT............... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................TGAGCTGGGGGATGCTGTGG............................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ...........................................................................................CACACCACATGCTCTGGCTATGT.................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTGAGCTGGGGGATGCTAGTGGGGA......................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ACACCACATGCTCTGGCTGAGA.................................................................. | 22 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGGGGGATGCTAGTGGGA.......................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........ACTGATCTTCAAGAGAAACTGAG..................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGGGGGATGCTAGTAT............................................................................................................ | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGTTTGGAACTGATCTTCAAGAGAAACTGAGTGACCAACACTACCTCTTGGTGAGCTGGGGGATGCTAGTGGGGAATGTCTTTGGGAAATCCACACCACATGCTCTGGCTAAGACTGTTTCTGTCACCAGGTGGGTGCTGGTGCCATTGGCTGTGAGATGCTCAAAGTCTTTGCCCTAGT .....................................................(((..(((.(((...(((.(((...((.....))..))).)))..))).)))..)))...................................................................... ..................................................51.................................................................118............................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR073955(GSM629281) total RNA. (blood) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR037905(GSM510441) brain_rep2. (brain) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | mjTestesWT3() Testes Data. (testes) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | GSM510444(GSM510444) brain_rep5. (brain) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | mjLiverWT3() Liver Data. (liver) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR059767(GSM562829) DN3_Dicer. (thymus) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (salivary gland) | SRR525244(SRA056111/SRX170320) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR037936(GSM510474) 293cand1. (cell line) | SRR525240(SRA056111/SRX170316) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR525241(SRA056111/SRX170317) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR037923(GSM510461) e9p5_rep1. (embryo) | SRR059765(GSM562827) DN3_control. (thymus) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR037901(GSM510437) testes_rep2. (testes) | SRR042478(GSM539870) mouse lung tissue [09-002]. (lung) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR065058(SRR065058) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR525243(SRA056111/SRX170319) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR037929(GSM510467) e7p5_rep3. (embryo) | GSM475281(GSM475281) total RNA. (testes) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR037904(GSM510440) brain_rep1. (brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......AACTGATCTTCAAGAGGC........................................................................................................................................................... | 18 | 8.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................GTCTTTGGGAAATCCACACCAC................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TCTTGGTGAGCTGGGGGAACAG................................................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................GTGGGTGCTGGTGCCAGG................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| .....................................ACACTACCTCTTGGTGAGCT........................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |